INTRODUCTION
The sika deer (Cervus nippon) is an important animal in China and has unique economic value. It is one of the main varieties used to produce high-quality velvet antlers, which are commonly used in traditional Chinese medicine [1]. However, due to overhunting and habitat fragmentation, the population of wild sika deer in northeastern China was on the verge of extinction by the end of the last century. In addition, sika deer have strong ornamental value because of their beauty, which could improve the potential for the development and utilization of tourism. Therefore, artificial rearing of sika deer has become the main method used to relieve the pressure of resource depletion and promote the development of tourism. Nevertheless, the diarrhea mortality rate of small artificially reared sika deer is greater, which causes great economic losses to the farming industry [2]. Hence, diarrhea has become the focus of prevention and control in artificially reared sika deer, although relevant research is still lacking.
Microbial colonization of the gastrointestinal tract (GIT) in neonatal ruminants is a key process that affects their health later in life [3]. Furthermore, the diversity and abundance of the microbial community in the GIT also markedly affect the feed efficiencies of ruminants [4]. The rumen microbiota provides 70% of the daily energy requirements of ruminants by fermenting undigested dietary substrates [3]. The microbiota of the mature rumen, comprising bacteria, archaea, fungi, and protozoa, can digest some forage feeds that cannot be broken down monogastrically and produce microbial crude protein (CP) and volatile fatty acids (VFAs), which serve as the main protein and energy sources for ruminants, respectively [3,5]; hence, previous studies have mainly focused on improving ruminant health and feed utilization efficiency by dietary intervention to regulate the GIT microbial community [6,7]. However, few studies have achieved lasting or consistent effects. This is mainly because a mature GIT microbial ecosystem has a highly stable host microbial community, and once manipulation is stopped, the composition of the microbial community will return to its original composition [8–10].
Generally, the GITs of newborn ruminants are considered sterile, and microbes quickly colonize the GIT after birth and gradually develop into a complex and stable microbiome [8], making it possible for the early life manipulation of the GIT microbiome to have lasting effects on adult ruminants. In ruminants, initial GIT microbial colonization is primarily determined by maternal-offspring microbial exchange through various methods, such as the exposure of neonates to a dam’s vagina, colostrum, breast milk, and skin [3]. Studies have shown that a lack of contact with adult animals and artificial milk feeding can limit rumen microbiota development and negatively affect feed digestibility [11,12]. Digestive disorders are the most common cause of diarrhea in small ruminants [3]. Therefore, compared with reared sika deer in dams, artificially reared sika deer are removed from their mothers immediately after birth and fed artificial milk, which may increase the likelihood of causing health and digestive problems.
In the present research, we hypothesized that repeated oral transplantation of maternal rumen microbiota could promote GIT microbiota community development and improve the health of sika deer calves. In this research, our objectives were to explore how to reconstruct the GIT microbial community of newborn sika deer through repeated oral transplantation of the microbiota in rumen fluid from a mother sika deer to her own offspring and to reveal the relationship between GIT colonization and development in sika deer.
MATERIALS AND METHODS
Healthy newborn sika deer (n = 12) and their mothers (n = 12) were selected from the Dongfeng County Sika Deer Industry Development Bureau in Liaoyuan, China. The separation of newborns from their mothers and milk distribution are the main methods for artificial rearing of sika deer. Therefore, 12 newborn sika deer were separated from their mothers immediately after birth under artificial rearing conditions (indoors). These 12 deer were randomly divided into control (CON; n = 6, half male and female) and fresh rumen fluid (FRF; n = 6, half male and female) groups, placed in two separate pens (2 m × 2 m) and acclimatized for three days (after birth, all the deer calves were fed approximately 800 mL of bovine colostrum divided into 5 doses for three consecutive days). The sika deer calves in the CON group did not receive any supplements, while those in the FRF group received 10 mL of FRF (added to the milk bottles) from their own mothers daily for four consecutive weeks. The sika deer calves in the CON and FRF groups were fed the same diet of commercial pasteurized pure milk (g/100 mL) containing 3.0 CP, 3.7 ether extract (EE), 4.8 carbohydrate, 0.062 sodium, and 0.10 calcium five times daily at 5:00, 9:00, 13:00, 17:00, and 21:00 h and had free access to commercial starter grain (31% corn, 44% soybean meal, 13% cooked beans, 9% wheat bran, and a 3% mixture of vitamins and mineral salts) on a dry matter (DM) basis: 98.35% organic matter (OM), 28.12% CP, 0.77% calcium, and 0.56% phosphorus and clean water. To meet the increasing nutritional needs of the newborn sika deer, the milk volume was increased from 1,000 mL/d on day 1 of the experiment (4 days old) to 1,850 mL/d on day 35 of the experiment (with the amount being increased by 25 mL per day). For the transplantation of microbiota from maternal rumen fluid, FRF samples were collected weekly (a total of 4 times) from each mother sika deer (after calving) corresponding to the deer calves in the FRF group via oral tubing [13]. Briefly, a flexible polyvinyl chloride tube with approximately 20 holes in the probe head was warmed with hot water and inserted through the esophagus into the rumen. Next, the rumen contents were obtained using an electric vacuum pump (Wertheim, Germany) connected to a sterile collection container. These samples were collected before the morning feeding and were filtered through cheesecloth (four layers) under a constant flow of CO2 to remove large feed particles (inocula) and then stored at 4°C in aliquots of 10 mL to be used for up to one week for inoculation. In addition, aliquots of 5 mL of rumen fluid samples from the mothers of sika deer calves in the CON and FRF groups were collected one time and immediately frozen at −80°C for DNA isolation (the mother samples were named MCON and MFRF, respectively). The mother sika deer were maintained in an individual outdoor pen and were fed with 40% commercial concentrate (67% corn, 20% soybean meal, 10% wheat bran, and a 3% mixture of vitamins and mineral salts) and 60% forage mixture (alfalfa hay: corn silage: dry oak leaves = 1:1:1), and the nutritional composition of the commercial concentrate (on a DM basis) was 98.90% OM, 15.81% CP, 0.76% calcium, and 0.58% phosphorus, while that of the forage mixture was 93.83% OM, 11.19% CP, 2.29% EE, and 36.05% crude fiber (CF). All animals had free access to clean water and food. Food was provided twice daily, at 8:00 and 16:00 h.
Ruminal development is divided into three stages: the nonrumination stage (from birth to 21 days), the transitional stage (from 21 to 56 days), and the rumination stage (from 56 days onward) [3]. The diarrhea incidence of young ruminants during the preruminant stage is higher; therefore, the experimental period of this study was set at the mid-transitional stage of 35 days. The body weight (BW) of sika deer calves was weighed weekly to calculate their average daily gains (ADGs). Fecal consistency was inspected by the farm workers five times daily (during milk feeding time) and scored on a scale from 1 to 4. Codes 1, 2, 3, and 4 were defined as normal consistency, semiformed or pasty, loose feces, and watery feces, respectively. Sika deer calves with a fecal score greater than or equal to 3 were considered positive for diarrhea [14]. Fecal samples were collected from the sika deer calves on experiment days 1 (before inoculation with maternal rumen microbiota), 28 (end of inoculation with maternal rumen microbiota), and 35 (one week after the end of the maternal rumen fluid inoculation period to ensure that the microorganisms involved in collected samples represent those that have colonized the GIT of sika deer calves and not from the inocula) and then quickly frozen at −80°C for DNA extraction. Moreover, on the final day of the experiment, blood samples and rumen fluid samples of each sika deer calf were collected. Blood samples were collected from the jugular vein, and serum was harvested by centrifugation (3,000 rpm and room temperature for 15 min) to determine aspartate aminotransferase (AST), alkaline phosphatase (ALP), and blood urea nitrogen (BUN) levels using commercial ELISA kits (Nanjing Jian Cheng Bioengineering, Nanjing, China). The data were measured by an autoanalyzer (Selectra-E, Clinical Data, Smithfield, RI, USA). Rumen fluid samples were withdrawn through orogastric intubation [13] and filtered through cheesecloth as previously described. An aliquot of 5 mL of rumen fluid from each sika deer calf was preserved at −80°C until DNA extraction. The pH values of the rumen fluid were measured immediately using a pH meter, and two subsamples were taken to determine the VFAs and ammonia-N concentrations using gas chromatography [15] and the colorimetric method [16], respectively. More details on method were provided in the Supplementary materials.
Metagenomic DNA was extracted from the rumen and fecal samples according to the manufacturer’s instructions. The quality of the DNA samples was assessed by 1.2% agarose (Invitrogen, Waltham, MA, USA) gel electrophoresis, and the concentrations were quantified by a Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen). The 16S rRNA gene hypervariable V3-V4 amplicon libraries were generated using 341F-CCTAYGGGRBGCASCAG and 806R-GGACTACNNGGGTATCTAAT primers [17]. The amplicon libraries were then sequenced with 2 × 250 paired-end sequencing on an Illumina MiSeq platform. The generated sequence data were processed and analyzed using Quantitative Insights Into Microbial Ecology 2 (QIIME2) (version 2019.4) [18]. Briefly, the DADA2 plugin was used to denoise the forward and reverse reads by filtering out low-quality reads with a Q-value of < 25 and to merge the reads before removing chimeras and singletons [19]. Finally, on average, 42,496 (35,501–50,716) and 49,005 (33,383–91,467) high-quality sequences resulted from each rumen fluid (Supplementary Table S1) and fecal sample (Supplementary Table S2), respectively, for further analysis. Amplicon sequence variants (ASVs) were classified into taxa based on the Greengenes database (version 13.8) [20] using a naive Bayes classifier [21]. Alpha diversity measurements, including the Chao1 index and Shannon index based on the number of observed unique ASVs were conducted using QIIME2 (version 2019.4) to compare the microbial community diversity with the single rumen fluid or fecal sample. Beta diversity based on Bray‒Curtis distance was calculated using the vegan package of R to compare the overall dissimilarity of microbiota between different groups in rumen fluid or fecal samples through analysis of similarity (ANOSIM) and shown by principal coordinates analysis (PCoA). Species composition differences were analyzed at the phylum, family, and genus levels to determine the differences in the abundance of microbial communities in the rumen fluid or fecal samples between the FRF and CON groups. In addition, the relative abundances of microbial taxa and the number of microbial taxa shared by and solely observed in the rumen and fecal samples were visualized using heatmaps and Venn diagrams, respectively [14]. To identify the cooccurrence of microbiota established in the GITs of sika deer calves and inocula, the taxa that differed significantly in abundance between the inocula (MFRF) and the rumen fluids of sika deer calves in the CON group were identified using linear discriminant analysis effect size (LEfSe) analysis conducted by LEfSe software with an LDA score > 2 [22]. The taxa that were more abundant in the MFRF samples than in the rumen samples from the sika deer calves in the CON group were referred to as “inoculum-predominant taxa” (biomarkers of inocula). The same comparison was made between the CON and FRF groups to infer the biomarkers that might be established by MRMT. Specifically, the taxa that had high abundances in both the inocula and the rumen fluid of the FRF sika deer calves but not in the rumen fluid of the CON sika deer calves were considered as probable biomarkers of the rumen microbiota of the mother deer donors. More details on method were provided in the Supplementary materials.
The number of animals (n) used in the experiments is listed in the Tables and Figures. Data were assessed in GraphPad Prism software (version 8) with the default parameter according to experience. The obtained data were subjected to normal distribution tests using the Shapiro‒Wilk test and Kolmogorov‒Smirnov test. Diarrheal status, serum biochemical parameters, animal performance, and rumen microbial fermentation data were analyzed by unpaired two-tailed Student’s t tests. Alpha diversity, Bray–Curtis distance, and microbial proportions (comparisons of deer calves between the FRF and CON groups) were analyzed using the nonparametric Mann‒Whitney test (between two groups) or Kruskal‒Wallis test (more than two groups). Dissimilarities in the ruminal or fecal microbial community were conducted by ANOSIM based on Bray‒Curtis distances with 999 permutations using the vegan package of R. LEfSe analysis was calculated by LEfSe software with an LDA score > 2. Pairwise comparisons were adjusted by false discovery rate. Data are presented as the mean ± SD, and differences were considered significant when the p-value was below 0.05 and trends when 0.05 < p < 0.10.
RESULTS
During the whole experimental period, one sika deer calf in the CON group died due to pneumonia on the 7th day of the experiment, and no samples were obtained from this individual. All the sika deer calves (5 of 5) in the CON group eventually developed diarrhea, and the average duration of diarrhea for each sika deer calf was 2.12 ± 0.88 days, while in the FRF group, most of the sika deer calves also developed diarrhea (4 of 6), but the average duration of diarrhea was markedly decreased (0.70 ± 0.25 days; p = 0.008; Table 1). The serum AST (p = 0.650), ALP (p = 0.937), and BUN (p = 0.604) levels were not markedly different between the sika deer calves in the CON and FRF groups (Supplementary Fig. S1). The initial body weights of the sika deer in the CON and FRF groups were 5.85 ± 0.44 kg and 6.64 ± 1.37 kg, respectively, and there was no significant difference (p = 0.292) in these weights between the two groups (Table 2), suggesting successful randomization when establishing comparable trial groups. The sika deer calves in the CON group gained 0.16 ± 0.03 kg of weight per day, which was not significant compared with that of the sika deer calves in the FRF (0.15 ± 0.02 kg/d) group (p = 0.471; Table 2). In the rumen fluid samples, the concentrations of acetic acid (p = 0.004), butyric acid (p = 0.006), valeric acid (p = 0.027), and total VFAs (p = 0.005) in the FRF group were significantly greater than those in the CON group; the pH value (p = 0.019) was markedly lower than that in the CON group; the ammonia nitrogen concentration (p = 0.152) of the rumen fluid samples did not differ between the CON and FRF groups (Table 3).
1) n, the number of sika deer calves; CON, control group sika deer calves inoculated without fresh rumen fluid and were grown separately from mother deer over the trial; FRF, fresh rumen fluid group sika deer calves inoculated with fresh rumen fluid and were grown separately from mother deer over the trial.
1) n, the number of sika deer calves; CON, control group sika deer calves inoculated without fresh rumen fluid and were grown separately from mother deer over the trial; FRF, fresh rumen fluid group sika deer calves inoculated with fresh rumen fluid and were grown separately from mother deer over the trial.
1) n, the number of sika deer calves; CON, control group sika deer calves inoculated without fresh rumen fluid and were grown separately from mother deer over the trial; FRF, fresh rumen fluid group sika deer calves inoculated with fresh rumen fluid and were grown separately from mother deer over the trial.
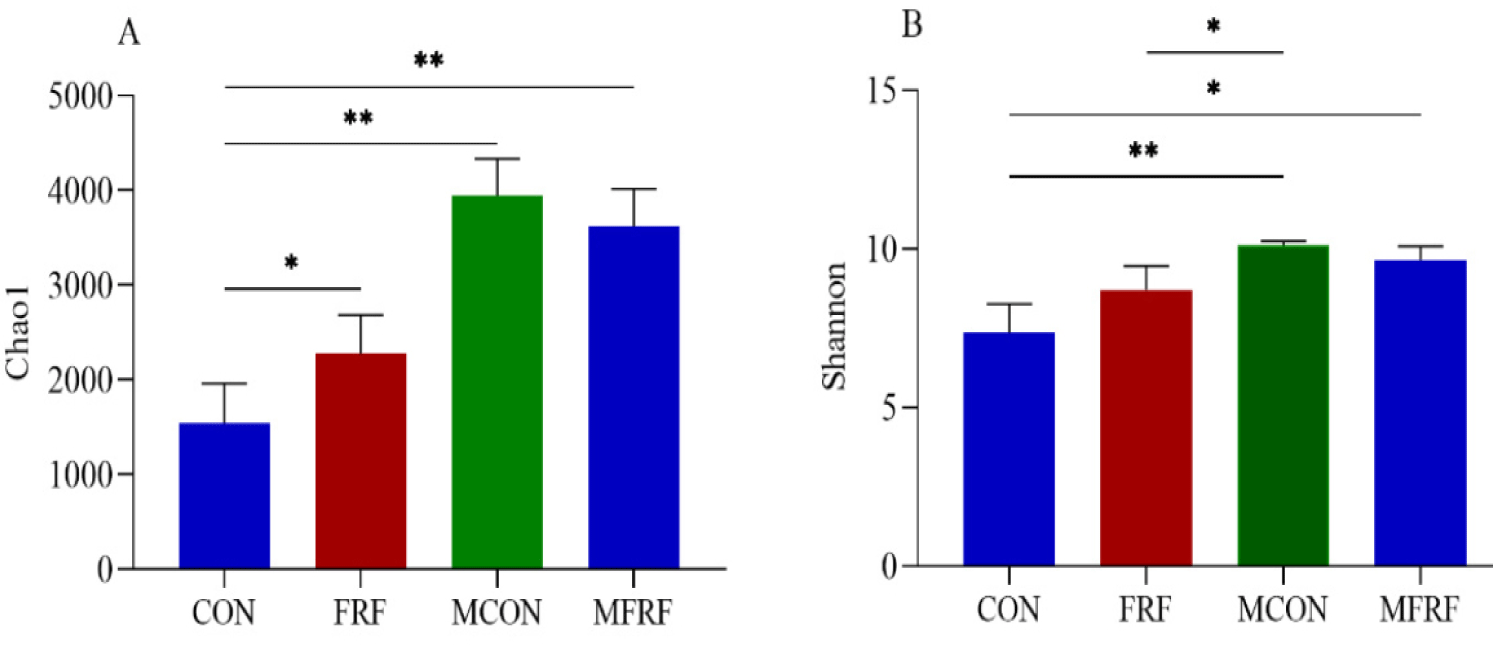
In total, 934,915 high-quality sequences were identified as 28,062 ASVs with an average Good’s coverage value of 98.16 ± 0.94% for all rumen fluid samples. The Chao1 index of the rumen microbiota in the FRF group was significantly higher than that in the CON group (p = 0.017); moreover, the rumen microbial communities in the samples from the mothers were more diverse and had higher Chao1 and Shannon indices than those in the samples from the sika deer calves (p < 0.05; Fig. 1). The PCoA scatter plot based on Bray‒Curtis distance (Fig. 2A) and the heatmap analysis of microbial taxa (Supplementary Fig. S2A) showed that there was high microbial community similarity among the maternal samples (ANOSIM, MCON vs. MFRF: p = 0.495) but that the maternal samples were different from the sika deer calf samples (ANOSIM, CON vs. MCON: p = 0.007; FRF vs. MFRF: p = 0.004); in addition, the ruminal microbial communities in the sika deer calves were significantly different between the calves in the CON and FRF groups (ANOSIM, CON vs. FRF: p = 0.002). The Bray‒Curtis distance between the FRF and MFRF groups was significantly lower than that between the CON and MCON groups (p < 0.0001; Fig. 2B). The Venn diagrams revealed that the calves in the FRF group shared more rumen microbial phyla, families, or genera with maternal samples than did the calves in the CON group (Supplementary Fig. S2B). These results suggested that MRMT caused the ruminal microbial communities of artificially reared sika deer calves to shift such that they resembled those of maternal samples. On day 35 of the experiment (Figs. 2C, 2D, and 2E and Supplementary Fig. S3), the relative abundances of microbial taxa, such as BS11 (p = 0.028) and Butyrivibrio (p = 0.045) were markedly higher in the ruminal microbiota of deer in the FRF group than in the CON group.
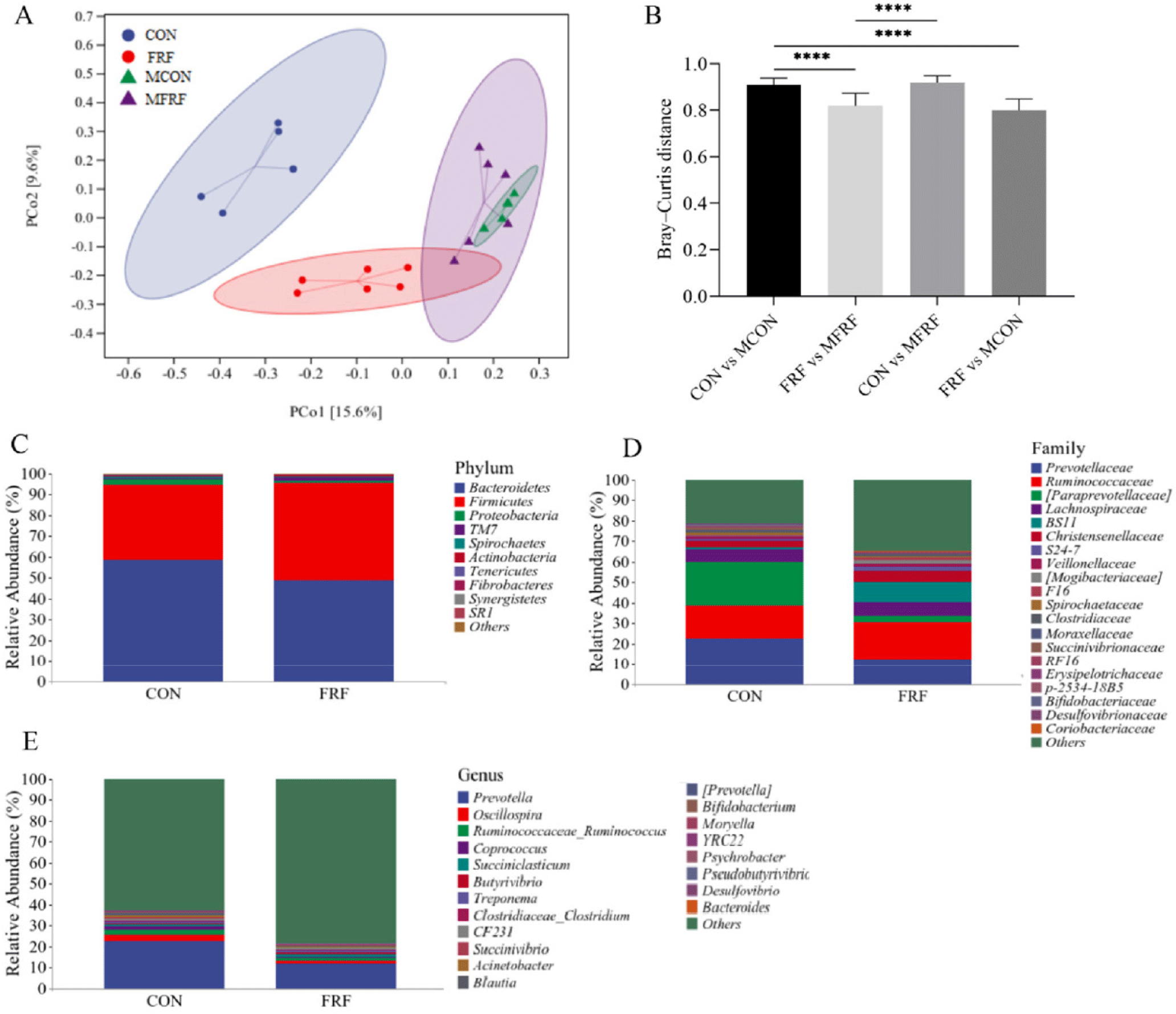
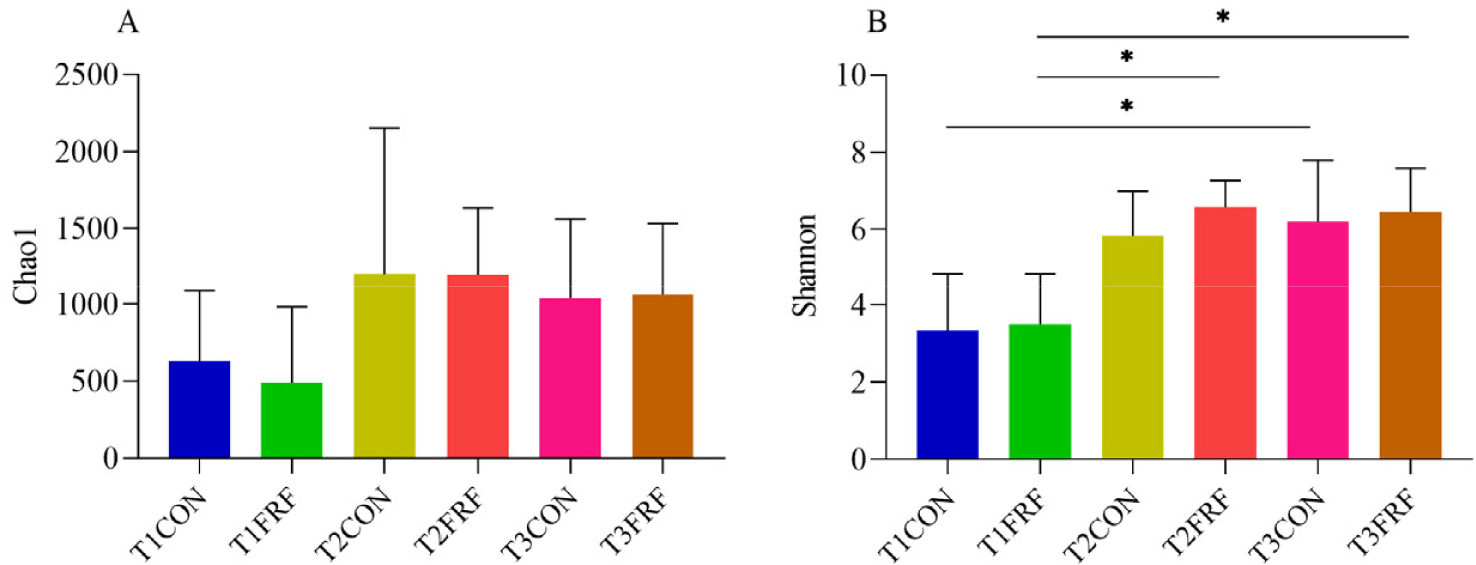
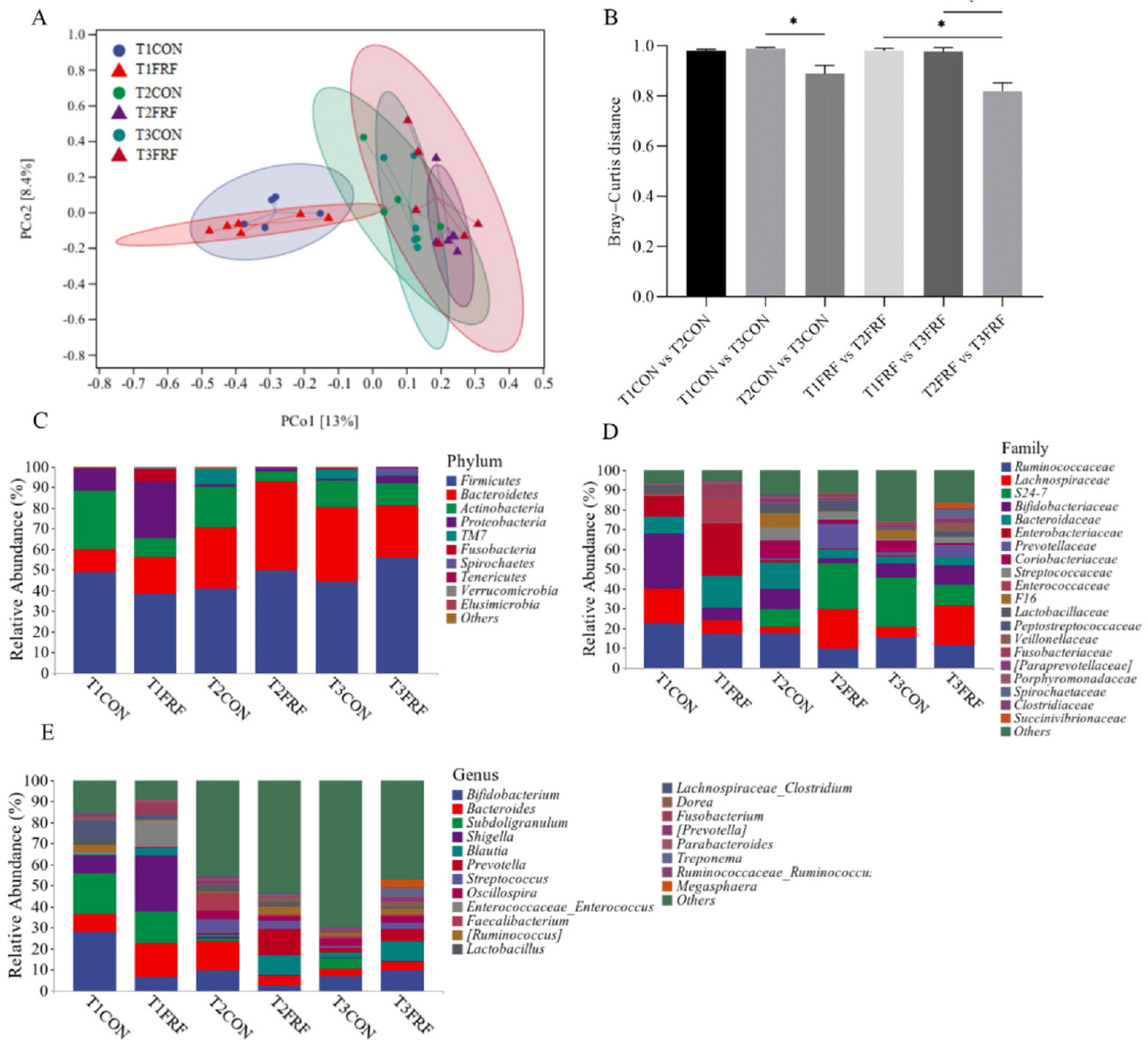
In total, 1,617,169 high-quality sequences were identified as 18,151 ASVs with an average Good’s coverage of 99.45 ± 0.44% for all the fecal samples. In the CON group, the Shannon index was significantly higher on day 35 than on day 1 of the experiment (p = 0.044), while in the FRF group, the Shannon index was significantly higher on days 28 (p = 0.024) and 35 (p = 0.021) than on day 1 of the experiment (Fig. 3). The PCoA scatter plot based on Bray‒Curtis distance (Figs. 4A and 4B) and heatmap analysis of microbial taxa (Supplementary Fig. S4A) showed that the microbial communities in the fecal samples of the CON and FRF groups were significantly different from day 1 of the experiment to days 28 (ANOSIM, T1CON vs. T2CON: p = 0.017; T1FRF vs. T2FRF: p = 0.002) and 35 (ANOSIM, T1CON vs. T3CON: p = 0.008; T1FRF vs. T3FRF: p = 0.003), but there was no significant difference between days 28 and 35 of the experiment (ANOSIM, T2CON vs. T3CON: p = 0.266; T2FRF vs. T3FRF: p = 0.424). The fecal microbial communities of the CON and FRF groups were very similar during the first day and final day of the experimental period (ANOSIM, T1CON vs. T1FRF: P = 0.638; T3CON vs. T3FRF: p = 0.290), but the Bray‒Curtis distance between 28 and 35 days of the experiment in the FRF group was significantly lower than that between 1 and 28 days (p = 0.039) and between 1 and 35 days (p = 0.026) of the experiment, while the Bray‒Curtis distance between 28 and 35 days of the experiment in the CON group was only significantly lower than that between 1 and 35 days of the experiment (p = 0.020). In addition, the Venn diagrams (Supplementary Fig. S4B) showed that on experiment day 35, the fecal samples of calves in the FRF group shared more microbial families or genera than did the fecal samples of calves in the CON group. The taxonomic composition analysis of the fecal microbiota (Figs. 4C, 4D, and 4E and Supplementary Fig. S5) showed that on experiment day 1, in the fecal samples of the sika deer calves, Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria were the predominant phyla, and Firmicutes was the most abundant; Ruminococcaceae, Lachnospiraceae, Bifidobacteriaceae, Bacteroidaceae, and Enterobacteriaceae were the most common families, and Bifidobacterium, Bacteroides, Subdoligranulum, and Shigella were the most common genera. Notably, on experiment day 1, the relative abundances of taxa in the FRF group were not significantly different from those in the CON group (p > 0.05), while on experiment day 35, the relative abundances of Lachnospiraceae (p = 0.017) and Butyrivibrio (p = 0.022) were markedly greater in the fecal microbiota of the FRF group than in that of the CON group.
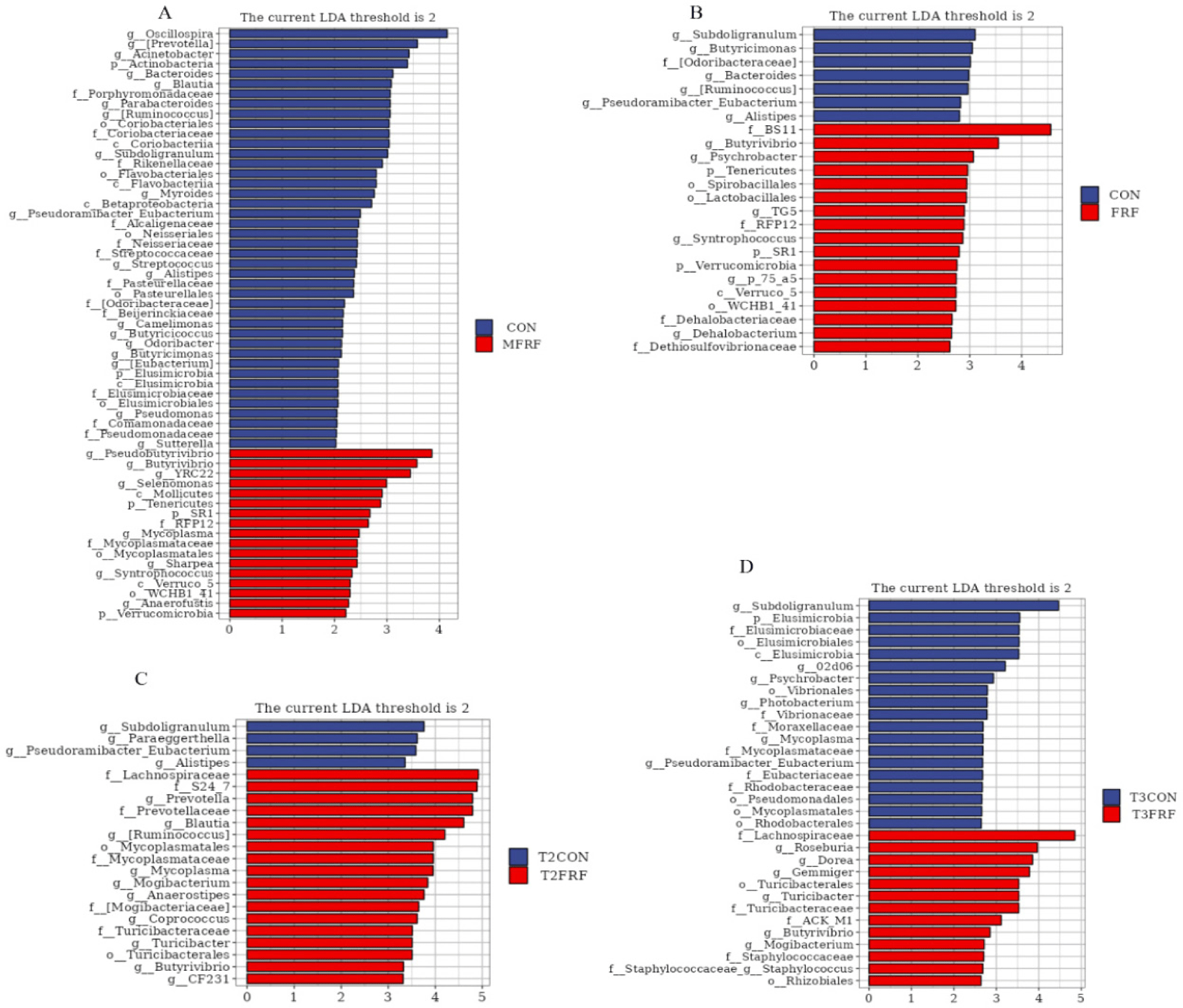
LEfSe analysis showed that 7 inoculum-dominant taxa were enriched in the rumens of sika deer calves in the FRF group (Figs. 5A and 5B), which were assigned to the genera Butyrivibrio, Tenericutes, RFP12, SR1, Verrucomicrobia, Verruco_5, and WCHB1_41. Moreover, the relative abundances of Lachnospiraceae, Turicibacter, Turicibacteraceae, Mogibacterium, and Butyrivibrio were significantly enriched in the fecal samples from the sika deer calves in the FRF group on experimental days 28 (Fig. 5C) and 35 (Fig. 5D). Among these taxa, Butyrivibrio is an inoculum-dominant taxon.
DISCUSSION
The diarrhea of artificially reared sika deer calves is related to the economic losses of the deer industry. Antibiotics are widely used for the treatment and prevention of common gastrointestinal diseases. Considering the adverse impacts of antibiotics on food security and the global ecological chain, alternatives to antibiotics are urgently needed in the livestock industry [23]. There is increasing evidence that mammalian host phenotypes can be manipulated by fecal microbiota transplantation, demonstrating that the microbiota of the GIT plays important roles in host health [24–26]. While the rumen microbiota shares many microorganisms with the fecal microbiota [27], the rumen microbiota has few or none of the pathogens that cause diarrhea in young ruminants, such as enterotoxigenic Escherichia coli, Clostridium perfringens, Cryptosporidium parvum, rotavirus, and coronavirus [14]. Moreover, the initial microbial colonization of the GIT in ruminants is primarily determined by maternal-offspring microbiota exchange [3]. Therefore, for ruminants, MRMT may also be useful in assessing the potential associations between GIT microbiota and host phenotypes. Our study demonstrated that MRMT significantly decreased the duration of diarrhea in sika deer calves. This result may be multifaceted, but inoculation with rumen fluid from adult ruminants has been shown to increase the activities of the intestinal immune system, and the development of microbiota in young ruminants has been demonstrated in earlier studies [28,29]. Therefore, MRMT may be an effective strategy to improve the gut health of sika deer in artificial rearing. To reveal the potential mechanism of MRMT and determine which GIT microbes have the potential to promote GIT development and antidiarrheal effects, in our research, we systematically studied the effects of MRMT on serum biochemical parameters, animal performance, rumen microbial fermentation, and the communities of microorganisms in the ruminal fluid and feces of sika deer calves.
In our study, the sika deer calves in the FRF group were given ruminal fluid daily for 28 days, which may have resulted in oxidative stress or toxicity. Therefore, several major indices suggestive of hepatotoxicity (AST and ALP) and nephrotoxicity (BUN) were examined. The levels of ALT, ALP and BUN were not significantly different between the CON and FRF groups. These results indicated that MRMT had no adverse effects on the organs of the sika deer calves, such as the liver and kidneys. No significant difference was observed in the ADGs of the CON and FRF groups, suggesting that although the ruminal microbiota composition can be altered during rumen development, these microbial changes do not necessarily lead to significant improvements in ruminant growth [30].
In newborn ruminants, the GIT is normally considered sterile, and the microbial community quickly establishes in the GIT after birth [3]. Once a mature microbial population has been established, the microbial community is usually believed to have reached a stable status. Stability refers to the existence of a stable equilibrium in the microbial community; once stability is reached, disturbances in the microbiota composition lead to changes, but the microbial community as a whole has the ability to restore these changes to its original status [31]. Generally, microbial communities with higher stability have stronger adaptability to external disturbances [3]. Therefore, young animals are more likely than adult animals to develop gastrointestinal diseases. Moreover, studies have reported that gut microbiomes with lower diversity are more susceptible to pathogens [32,33]. In the current study, we observed higher alpha diversities of the rumen microbiota in the sika deer calves in the FRF group than in the CON group. Furthermore, the Bray‒Curtis distance was lower and the number of taxa shared was higher between the FRF and MFRF groups than between the CON and MCON groups, suggesting that MRMT increased rumen microbiota diversity and may have shortened the time needed for the rumen microbiota to reach stability in the artificially reared sika deer calves. There were no significant differences between the CON and FRF groups in the microbial diversities of the fecal samples throughout the trial period, but the alpha diversities in the CON group were higher on day 35 than on day 1 of the experiment, while those in the FRF group were higher on days 28 and 35 than on day 1 of the experiment; moreover, the Bray‒Curtis distance in the FRF group observed from days 28 to 35 of the experiment was markedly lower than that between 1 and 28 days and between 1 and 35 days of the experiment; however, in the CON group, the Bray‒Curtis distance observed from days 28 to 35 of the experiment was only significantly lower than that between 1 and 35 days of the experiment. These data suggested that although the temporal changes in the fecal microbiota diversity of the sika deer calves in the CON group were similar to those in the FRF group, MRMT improved the temporal development of gut microbial diversity and stability from days 1 to 28 of the experiment in the sika deer calves in the FRF group. These findings demonstrated that MRMT may be an effective method to restore the GIT microbiota of artificially reared sika deer calves.
In the present study, Firmicutes was the most abundant in the feces of these sika deer calves, which is consistent with the results from the feces of preweaned dairy calves [34]. The sequences from Bifidobacterium, Bacteroides, Subdoligranulum, and Shigella were the dominant microbiota of the deer calves on day 1 of the experiment. Escherichia–Shigella and Bacteroides are also abundant in the feces of the calf and colon of sika deer after birth [35,36]. The shared distribution of these microbiota from different ruminants indicates their general importance in the early growth period. Notably, Bifidobacterium and Subdoligranulum were comparatively abundant in the fecal samples of newborns. Bifidobacterium is a common probiotic that has been reported to be relatively abundant in fresh colostrum [37]. Therefore, it may be transferred from bovine colostrum that is fed to deer calves. Moreover, Subdoligranulum has been demonstrated to be positively correlated with gut health. These results proved that all the deer calves had good health status at the start of the experiment.
To understand the effects of MRMT on key phylotypes of the GIT microbiota in sika deer calves, the microbial compositions of the rumen and feces of the sika deer calves in the CON and FRF groups were compared. Earlier studies reported that VFAs can be absorbed by rumen epithelial cells as additional energy for the host [38,39]. BS11 was associated with higher rates of VFA (acetate and butyrate) production in the rumen [40]. Lachnospiraceae and Butyrivibrio are butyrate-producing bacteria and play an important function in cellulose degradation, maintenance of health and productivity of ruminants [41–43]. In the present study, the relative abundances of BS11 and Butyrivibrio were higher in the rumens of FRF group calves than in those of CON group calves. As a result, calves in the FRF group had greater rumen fermentation ability (higher concentrations of acetic acid, butyric acid, and total VFAs) than calves in the CON group. Moreover, the relative abundances of Lachnospiraceae and Butyrivibrio were higher in the feces of FRF group calves than in those of CON group calves. Therefore, MRMT may have the potential to improve the degradation of structural polysaccharides in the GITs of sika deer calves and play an important role in promoting the maturation of the GIT immune system in ruminants. In addition, given that transferring the rumen microbiota from sika deer mothers to their offspring significantly decreased the occurrence of diarrhea among the sika deer calves, we mainly focused on the cooccurrence of the microbiota established in the GITs of the sika deer calves and the microbiota in the inocula to screen potential biomarkers related to diarrhea resistance. Seven inoculum-predominant taxa, Butyrivibrio, Tenericutes, RFP12, SR1, Verrucomicrobia, Verruco_5, and WCHB1_41, were more abundant in the rumens of sika deer calves in the FRF group than in those in the CON group, suggesting that these microorganisms might be transferred from maternal rumens to the rumens of artificially reared sika deer calves. Although these microorganisms are not the dominant microbiota in the ruminal microbial community, they may play an important role in the main functions of the rumen microbial community, for instance, dealing with certain secondary metabolites to maintain host health. Earlier studies of the gut microbiome were largely limited to identifying the most abundant microbiota associated with health or disease; however, the metabolic functions performed by low-abundance microbiota may also control gut balance. Research has reported that Actinobacteria are relatively rare in the healthy human gut but are positively associated with gut microbiota diversity, play a key role in the biodegradation of complex starches, and may be involved in suppressing dysbiosis in patients with inflammatory bowel disease [44,45]. Therefore, the importance of these low-abundance species cannot be ignored; however, the correlation between these candidate microorganisms and host health needs to be further studied. Furthermore, the genus Butyrivibrio was the only inoculum-dominant taxon that was also more abundant in the fecal samples of sika deer calves in the FRF group than in the samples from the CON group on experimental days 28 and 35. Thus, Butyrivibrio may be a potential target of MRMT. However, fewer inoculum-dominant genera were found in the feces than in the rumens of the artificially reared sika deer calves, suggesting that the rumen is more accepting of rumen microbial inoculation than the intestinal tract. The genus Butyrivibrio has been shown to effectively utilize plant polysaccharides, such as hemicellulose and pectin, which other bacteria cannot degrade to produce butyric acid [41]. Butyric acid is an important VFA produced by GIT microbial fermentation and can maintain gut health by regulating the immune system and/or maintaining the gut epithelial barrier [46,47]. Butyric acid also plays an important role in promoting rumen development, as mentioned above. Therefore, MRMT has the potential to promote GIT development in artificially reared sika deer calves.
CONCLUSION
Oral inoculation of artificially reared sika deer calves with rumen microbiota from their own mothers may substantially affect the establishment of the microbial community in the GIT; for example, it caused the ruminal microbial compositions of sika deer calves to resemble those of their mothers and promoted the temporal development of gut microbial diversity in sika deer calves from day 1 to day 28 of the experiment. Oral inoculation also increased ruminal fermentation and reduced the duration of diarrhea in sika deer calves. The genus Butyrivibrio was significantly enriched in both the rumen and feces of the sika deer calves after inoculation, indicating that this genus may be a potential target of MRMT. Taken together, these results reveal that MRMT may be a useful approach to redirect the development of microbiota in the GIT and improve the health of artificially reared sika deer calves.
















