INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs with a length of 22 nucleotides [1]. They regulate the transcription and post-transcription of gene expression by mRNA degradation or translational repression depending on the binding to the 3’ untranslated region (UTR) of target mRNA [2,3]. MiRNAs derived from various cell types are secreted into the extracellular space through exosomes, protein complexes, etc. [4], and then transferred to the body fluid. These are commonly known as circulating miRNA, showing potential as biomarkers for disease diagnosis [5]. While most studies on circulating miRNAs have focused on human and livestock blood [6,7], recent studies have identified miRNAs in saliva, leading to an increase in saliva-based research [8,9]. Saliva shares various physiological characteristics and allows for repeated specimen collection, similar to blood. Saliva contains miRNAs, proteins, and hormones, providing insights into its physiological properties and offering further understanding of several biological processes [10]. Blood collection is painful and stressful, whereas saliva sampling does not cause pain and can be easily performed even by untrained individuals [10,11]. Investigating salivary miRNAs provides promising insights for the non-invasive monitoring of health status and related diseases. Additionally, saliva samples may contain diverse microorganisms that can provide further insights into its biological characteristics and relationship with the gut microbiome. Therefore, saliva sampling has attracted considerable interest in human research [8,12,13]. The miRNAs and the microbiome are widely recognized as key indicators of livestock health, productivity, stress, and disease [14–16]. And miRNAs have been used as biomarkers for monitoring health conditions, including inflammation, metabolic disorders, and stress responses, while the microbiome offers insights into gut health, immune function, and overall well-being. In pigs, salivary miRNAs such as miR-19b, miR-27b, and miR-365 have been identified as potential biomarkers for assessing pain and stress, particularly in response to procedures like castration and tail docking, with their expression levels suggesting a viable approach for non-invasive pain monitoring [9]. However, studies on salivary circulating miRNA expression in pigs remain limited.
Therefore, this study aimed to screen salivary miRNAs in pigs and investigate their biological functions. Additionally, we aimed to preliminarily explore the presence and potential role of the salivary microbiome, thereby enhancing our understanding of the biological value of porcine saliva.
MATERIALS AND METHODS
Saliva samples were collected using specialized salivary tubes (Salivette, Sarstedt) from two adult female pigs, consisting of a gilt (Duroc, approximately 46-week-old) and a sow (Landrace, approximately 197-week-old). All animals were housed and cared for at the Kongju National University Animal Farm. And the cotton roll was kept in the pig mouth, allowing the animal to chew it for 2–3 min followed by centrifugation of sponge-containing Salivette tubes at 1,660×g for 20 min to collect saliva.
Small RNA was extracted from the saliva using a XENOPURETM Plasma/Serum Small RNA Purification Kit (Xenohelix). RNA quality was assessed using an Agilent Technologies TapeStation 2200 (Agilent Technologies). Sequencing was performed using 100 base single-end reads on a NovaSeq 6000 sequencer (Illumina).
Raw reads were quality-checked using FASTQC (v0.12.1) [17], and adaptor and low-quality sequences were trimmed using Cutadapt (v4.7) [18]. Trimmed reads (≥ 15nt) were then aligned to the porcine reference genome (Sus scrofa 11.1.106, GCA_000003025.6) using Bowtie1 (v1.3.1) [19] for detecting miRNAs. Novel and known miRNAs were predicted using miRDeep2 with miR databases for various species, including Sus scrofa, Bos taurus, Equus caballus, Ovis aries, Canis familiaris, and Homo sapiens [20]. Separately from this, the trimmed reads were also aligned using STAR (v2.7.11b) to the presence of other types of small RNA transcripts with splicing events in porcine saliva [21].
To further explore the potential biological functions and processes of miRNAs in porcine saliva, the potential target genes of miRNAs were predicted using the TargetScan (v8.0) tool [22]. For the predicted target genes of salivary miRNAs, the biological process, cellular component, and molecular function from gene ontology (GO) were used to annotate functions using the Database for Annotation, Visualization, and Integrated Discovery [23] using the default options. A GO bubble plot was generated using the SRplot tool [24].
RESULTS AND DISCUSSION
Saliva was collected from adult female pigs with well-developed salivary glands, and 26 known miRNAs were detected via small RNA sequencing (Fig. 1A and Table 1). Among these, miR-19b has been highlighted in a previous study as a potential biomarker of inflammatory responses in piglets after tail docking and castration [9]. Additionally, 223 novel miRNAs were identified based on miRNA sequence databases of other species. This result may be related to the relatively low number of annotated miRNAs in pigs in miRBase 22.1 [27], compared with that in humans and other livestock species [28], which likely explains the high number of novel miRNAs annotated. Alternatively, these novel miRNAs could be saliva-specific. The sizable detection of novel miRNAs supports the potential of saliva as a specimen for investigating miRNA profiles and their biological processes in pigs. Saliva is a body fluid sample that can be easily collected by anyone through a noninvasive method, and it has various molecular characteristics, which may provide valuable insight as potential biomarkers for disease diagnosis and evaluating individual disease resilience.

GO analysis of the target genes of known miRNAs revealed their involvement in key biological processes, such as transcriptional regulation, cell structure maintenance, and neural development, primarily functioning within the nucleus and cytoplasm (Fig. 1B). In particular, RNA polymerase II is the key enzyme responsible for protein-coding genes transcription [29], and miRNAs can regulate gene expression in both positive and negative ways [30]. Their interactions with RNA polymerase II suggest that miRNAs may act as significant regulators of gene expression. The cytoskeleton organization process is crucial for maintaining the structural stability of cells. MiRNAs involved in this process can regulate the expression of genes that control the assembly and dynamics of the cytoskeleton. According to previous studies, miRNAs are an important role in regulating cytoskeletal proteins [31–33], which affects cell signaling and structural integrity. Furthermore, the dentate gyrus development term, which is essential for neurogenesis and neuronal development [34], was significantly enriched in the GO analysis. This implies that miRNAs can regulate the expression of genes involved in dentate gyrus development [35]. In addition, the high enrichment of ‘protein binding’ suggests that salivary miRNAs may influence various physiological functions in pigs by regulating protein-protein interactions. These results suggest that porcine saliva contains functional miRNAs with important roles in the physiological regulation of gene expression and various physiological functions in pigs.
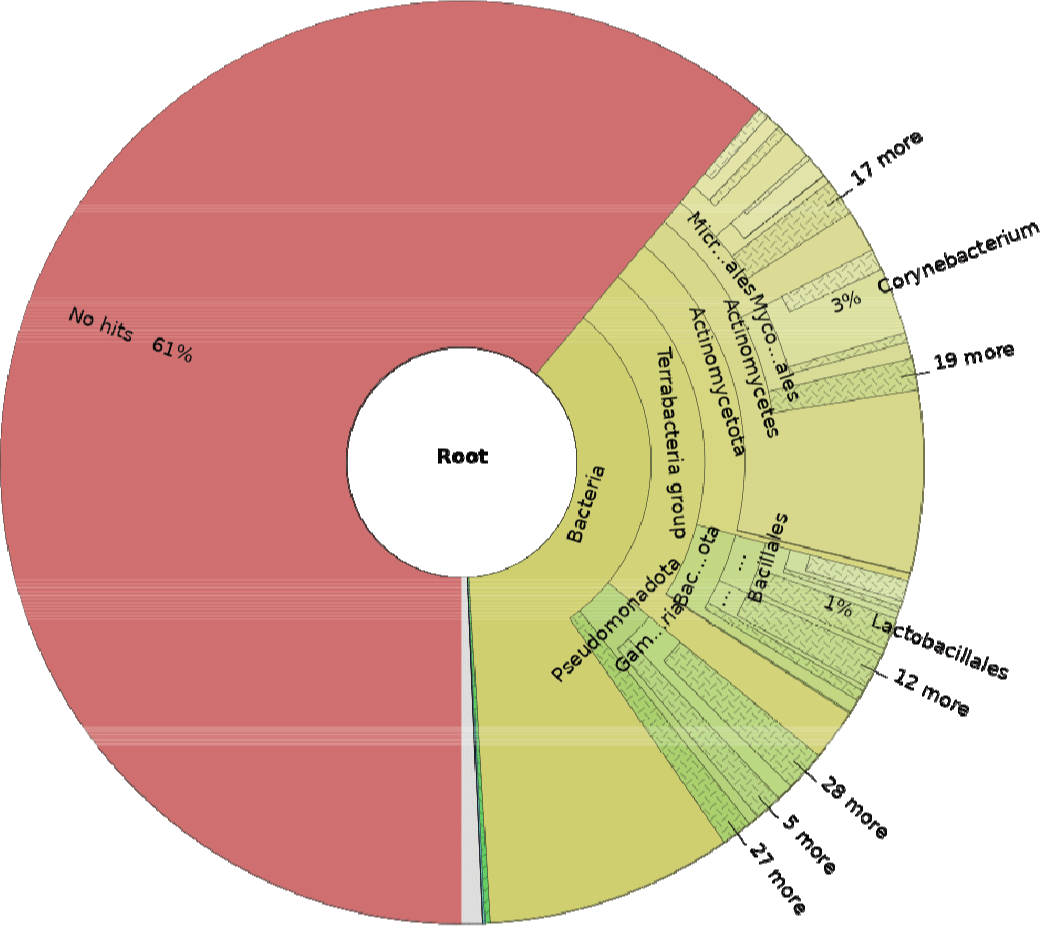
The STAR (29.4%–33.5%) and Bowtie (2.04%–3.42%) mapping rates differed notably (Table 2), likely due to ability of STAR to recognize splicing events [21]. Overall, the relatively low mapping rates suggest the substantial presence of externally derived microorganisms in porcine saliva. Classification via comparison with a diverse microbial species database identified approximately 39% of the total reads as microbes (Fig. 2), including the Corynebacterium genus, which is highly abundant in the oral microbiome [36]. Corynebacterium is a meaningful genus in the oral microbiome, particularly abundant in human saliva, where it contributes to maintaining oral health. Several studies have shown that Corynebacterium plays a role in restoring the balance of oral biofilms, suggesting its potential to promote oral health [37–39]. Additionally, they have been found to secrete various fatty acids with anti-inflammatory effects [40], further supporting their beneficial roles in regulating oral health. Overall, Corynebacterium appears to be strongly associated with oral health and may actively coordinate health-promoting activities. Recently, research has also identified Corynebacterium as a predominant genus in the oral microbiome of sows, suggesting its importance in maintaining the health of the oral microbiome in pigs [41]. This finding asserts the potential of Corynebacterium in influencing the overall health and resilience of pigs, particularly in relation to oral health and immune functions. Moreover, a reciprocal interaction between miRNAs and microorganisms have been extensively studied [42]. MiRNAs can regulate immune responses, which in turn may influence the abundance of the microbiome, including the abundance of beneficial bacteria [43]. Conversely, microorganisms can also affect host miRNA expression, influencing biological processes such as inflammation and immune responses [44].
| Bowtie (%) | STAR (%) | |
|---|---|---|
| Landrace | 2.04 | 29.4 |
| Duroc | 3.42 | 33.5 |
This result demonstrates the potential of saliva as a meaningful sample in pig microbiome research. Salivary RNA and microbiome composition can be influenced by various factors, such as diet [45], sample collection timepoint and methods [46]. Therefore, these variables could be considerable in further studies to improve the accuracy and reliability of saliva samples.
CONCLUSIONS
In this study, we identified both known and novel miRNAs in porcine saliva, highlighting its potential as a valuable sample for miRNA exploration and microbiome analysis. The diverse microbial presence, including known oral microbiota, such as Corynebacterium, suggests that saliva is a promising sample for noninvasive monitoring of microbiome diversity and physiological states in pigs. However, the origin of the unmapped reads remains unclear and requires further investigation. This study lays the groundwork for future research on the diagnostic potential of miRNAs and unidentified components in porcine saliva, offering new possibilities for noninvasive health monitoring in pigs.