INTRODUCTION
The porcine respiratory disease complex (PRDC) causes economic losses in the swine industry by reducing pork production efficiency and increasing feed costs, carcass disposal costs, and medical expenses [1]. Various factors, including overcrowded rearing conditions, porcine circovirus type 2 (PCV2), porcine reproductive and respiratory syndrome virus, swine influenza virus, Mycoplasma hyopneumoniae, and Pasteurella multocida influence the onset of PRDC [2]. Infections by PRDC-related viruses lead to proinflammatory cytokine expression and tissue damage in porcine lungs [3], which can be alleviated by anti-inflammatory drugs (AIDs). However, the development and selection of suitable AIDs for PRDC are limited because of a lack of research [4]. Therefore, in this study, we aimed to develop an AID for porcine.
Inflammation caused by infection is primarily mediated by macrophages. PCV2 targets macrophage populations, including alveolar macrophages (AMs), and induces a strong inflammatory response [5]. Increased expression of NOX2, which mediates reactive oxygen species (ROS) production, has been reported in PCV2-infected macrophages [6]. This induces autophagy and PCV2 replication, which can be inhibited by blocking autophagy or ROS [7,8]. Additionally, PCV2-infected macrophages show increased expression of proinflammatory cytokines, including tumor necrosis factor-α (TNFα) and cyclooxygenase-2 (COX2) [9]. The acute inflammatory response in macrophages is mediated by P2Y purinoceptor 14 (P2Y14), which is a member of the pyrimidinergic G protein-coupled receptor family. Activation of P2Y14 by uridine-5′-diphosphoglucose (UDPG), which is produced and secreted during glycogenesis, induces the expression of signal transducer and activator of transcription 1 (STAT1) and TNFα [10]. Although the ability of P2Y14 to recognize UDPG has been reported in porcine coronary arteries [11], the role of P2Y14 in porcine macrophages (PAMs) is unclear.
With the global ban on antibiotics in animal feed, there has been increased attention on developing eco-friendly antibacterial and anti-inflammatory strategies to maintain porcine health and productivity [12]. Succulent plants, which are also used as porcine feed, have shown some anti-inflammatory effects in porcine cells and are emerging as natural anti-inflammatory agents [13–15]. The succulent Aporocactus flagelliformis, also known as rattail cactus, is traditionally used in Mexico to treat heart disease and diabetes [16,17]. Inhibition of P2Y14 in porcine shows potential for treating these conditions [11,18]. Here, we focused on the therapeutic effects of A. flagelliformis on P2Y14 and various diseases.
In this study, we developed an A. flagelliformis water extract (AFWE) and investigated its anti-inflammatory effects on 3D4/31-PAMs, including ROS production, proinflammatory cytokine expression, and bactericidal activity. We also evaluated the effects of AFWE on P2Y14 metabolism. Limonin was identified using liquid chromatography-mass spectrometry (LC-MS) as a major compound of AFWE. The binding potential of limonin to porcine P2Y14 was assessed and the therapeutic effects of limonin on UDPG-induced inflammation were evaluated.
MATERIALS AND METHODS
Fresh A. flagelliformis (Xplant) was cut into 2–3 cm lengths, washed with deionized water (dH2O), and extracted with dH2O (300 mL dH2O for 100 g A. flagelliformis) for 15 min at 110°C using a WAC-60 autoclave (Daihan Scientific). The aqueous phase was collected, sterile filtered using a 0.2 μm cellulose-acetate filter (16534-K, Minisart, Sartorius), and stored at –80°C before use.
3D4/31 porcine alveolar macrophages (3D4/31-PAMs; ATCC-CRL-2844; ATCC) and A549 human alveolar epithelial cells (A549-AECs; CCL-185, ATCC) were maintained in a 5% CO2 atmosphere at 36.5°C. Cells were grown in a 4:6 ratio of Dulbecco’s modified Eagle’s medium (10-013-CVR, Corning) and Roswell Park Memorial Institute (RPMI) 1640 medium (10-040-CVR, Corning) supplemented with 10% (v/v) fetal bovine serum (TMS-013, Merck Millipore) and 1% (v/v) penicillin-streptomycin (LS202-02, Welgene).
Cells were cultured for 12 h prior to treatment. Cells were treated for 24 h, refreshed medium/treatment, and stimulated with 2 nM phorbol myristate acetate (PMA; P1585-1MG, Sigma-Aldrich). The treatments included 60 μg/mL AFWE, 30 μM limonin (A10531, Adooq Bioscience), and 200 µM UDPG (U4625-25MG, Sigma-Aldrich). Limonin was prepared at a final concentration of 50 mM in 99.9% dimethyl sulfoxide (DMSO; sterile, cell culture grade).
To quantify cell viability, cells were cultured with 10% (v/v) water-soluble tetrazolium salt-8 (WST8) reagent (QM2500, BIOMAX) for 2 h, and the optical density at 450 nm (OD450) was measured using a FilterMax F3 microplate reader (Molecular Devices). For quantification of proliferation, cells were harvested and stained with 0.2% (v/v) trypan blue (15250-061, Gibco) for 1 min, and viable cells were counted using a hemocytometer.
Cells were cultured with 1 µM 2′,7′-dichlorofluorescein diacetate (H2DCFDA; 35845, Sigma-Aldrich) for 30 min, washed with phosphate-buffered saline (PBS; pH 7.4), harvested, washed with PBS, and analyzed by flow cytometry.
Cells were lysed in radioimmunoprecipitation assay buffer containing 1 mM phenylmethanesulfonyl fluoride (P7626-5G, Sigma-Aldrich) for 1 h at 4°C. The supernatant was collected by centrifugation at 14,000 RCF for 15 min. Protein concentration was quantified using Bradford’s assay with a bovine serum albumin (BSA; 10735086001, Roche) standard. Proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes (3010040001, Roche) using a HorizeBLOT 2M transfer system (ATTO). Membranes were blocked with 5% (w/v) skim milk, washed with tris-buffered saline [TBST; pH 7.6, 0.05% (v/v) Tween 20], and probed with primary antibodies at 4°C for 12 h. Membranes were then washed with TBST, proved with secondary antibodies, and washed with TBST. The membranes were then exposed to an enhanced chemiluminescence reagent, visualized using an X-ray film (EA8EC, Agfa-Gevaert), and quantified by densitometry analysis using ImageJ Ver. 1.5.3q (National Institutes of Health). The antibodies used for immunoblotting are listed in Table 1.
All procedures were performed according to the manufacturer’s instructions. RNA was isolated using TRIzol reagent (15596026, Invitrogen), quantified using NanoDrop, and converted to cDNA at a concentration of 1 μg using Oligo dT20 primers with the WizScript cDNA Synthesis Kit (W2202, Wizbiosolutions). cDNA was quantified using SYBR Green qPCR Master Mix (DQ485; BioFACT), StepOnePlus RT-PCR System (Applied Biosystems), and StepOne software Ver. 2.3. The fold change in mRNA expression was normalized to ribosomal protein S29 (RPS29) using the 2(− ΔΔCt) method. The primer sequences used for qRT-PCR are listed in Table 2.
3D4/31-PAMs (1×106 cells), Escherichia coli DH5α (1×107 CFU, colony forming unit), and 5% (v/v) porcine serum were mixed in a final volume of 1 mL of Hanks’ balanced salt solution (HBSS, pH 7.4) and incubated at 37°C for 1 h with shaking (180 rpm). After centrifugation at 12,000 RCF for 1 min, the supernatant (non-engulfed bacteria) was spread onto Luria-Bertani (LB; 244602, Becton Dickinson) agar plates. The pelleted cells (with engulfed bacteria) were washed twice with HBSS, suspended in 1 mL of RPM1640 medium, and incubated for 0, 20, and 40 min at 37°C with shaking (180 rpm). After each incubation period, the cells were lysed with dH2O for 5 min and spread on LB agar plates. The CFUs were counted after incubation for 12 h at 37°C. Images of the LB agar plates were captured using iPhone X (Apple).
Cells were cultured with 1 µg/mL acridine orange (A6014, Sigma-Aldrich) for 15 min, washed twice with PBS, harvested, washed with PBS, and analyzed by flow cytometry.
Cells were suspended in PBS containing 50 µM 2-NBD-glucose (2-NBDG; 11046-10MG, Cayman Chemical) and 0.1% (w/v) BSA, incubated for 30 min, washed with PBS, and analyzed by flow cytometry.
Cells were cultured with 1 µM BODIPY493/503 (D3922, Invitrogen) for 30 min, washed twice with PBS, and subjected to fluorescence microscopy or flow cytometry. For flow cytometry, cells were harvested, washed with PBS, and analyzed.
Glycogens were visualized using Best’s carmine staining [19] with minor modifications. Cells were fixed for 15 min with 3.8% (w/v) formaldehyde, washed with PBS, and stained with 0.625% (w/v) Best’s carmine solution (C1022; Sigma-Aldrich) for 30 min. Cells were then rinsed twice with dH2O containing 2% (v/v) methanol and 4% (v/v) ethanol, washed with ethanol for 1 min, and imaged. Glycogen content was quantified using the anthrone method [20] with minor modifications. Cells (1×105) were lysed in 50 μL of 30% (w/v) KOH for 20 min at 100°C and 350 μL of 43% (v/v) ethanol was added. The cell lysate (50 μL) was reacted with 100 μL of 0.2% (w/v) anthrone (319899, Sigma-Aldrich) at 100°C for 20 min. The OD620 of the lysate and glucose (G8270, Sigma-Aldrich) standards were measured using a FilterMaxF3 microplate reader.
Serial fractionation of AFWE was performed using ethyl acetate, ethyl ether, ethanol, and isopropyl ether (extra-pure grade). The AFWE was shaken in a specific solvent system for 10 min and allowed to stand at 25°C until the mixture formed two layers (1–2 h). The organic layer was then concentrated to 20 × in DMSO using a rotary evaporator.
LC was carried out using an Acquity UPLC system (Waters) with an Acquity BEH C18 1.7 μm column (2.1 × 100 mm). The LC processed the samples at 0.2 mL/min using water/methanol with 0.1% (v/v) formic acid at 40°C. MS was performed using the SYNAPT G2 platform (Waters). Molecules were identified using m/z CLOUD (https://www.mzcloud.org) and molecular structures were illustrated using ChemDraw Ultra Ver. 12.0.2 (CambridgeSoft).
The 3D structure of porcine-P2Y14 (AF-F1SJN3-F1) was downloaded from the AlphaFold Protein Structure Database Ver. 2022-11-01 (https://alphafold.ebi.ac.uk). The canonical SMILES of the ligands (Limonin #179651 and UDPG #8629, PubChem release 2021.10.14) were retrieved from the PubChem Database (https://pubchem.ncbi.nlm.nih.gov). Molecular docking and visualization were performed using DiffDock-L [21] hosted at Neurosnap.ai (https://neurosnap.ai).
Flow cytometry was performed using a Guava easyCyte Flow Cytometer (Merck Millipore) and Guava InCyte software Ver. 2.6. An average of 3×103 cells was measured for the single-channel assay and an average of 5×103 cells was measured for the dual-channel assay. Flow cytometry plots and fluorescence intensities were obtained using the FlowJo Ver. 10.6.2 (TreeStar).
Microscopy was performed using DMi8 fluorescence microscope (Leica Camera). LAS X software Ver. 2.0.0.14332 was used for the fluorescence images, and LAS software Ver. 4.7.1 for the bright-field images. To display representative images, contrast and brightness adjustments were processed using Photoshop 2024 (Adobe).
Statistical analyses were based on at least 3 independent biological experiments and were performed using GraphPad PRISM software Ver. 10.2.3 (GraphPad). All data are shown as mean ± SD. Analysis of variance (ANOVA) with Tukey’s multiple comparison test or unpaired two-tailed Student’s t-test was used for statistical analyses. Statistical significance was set at p < 0.05.
RESULTS
To determine the anti-inflammatory properties of A. flagelliformis, we developed AFWE and evaluated its bioactivity against ROS production, cytokine expression, and bactericidal activity in 3D4/31-PAMs. First, fresh A. flagelliformis was extracted using a water extraction method (Fig. 1A and 1B). The dry weight of AFWE was found to be 6 mg/mL, giving an extraction efficiency of 1.8% based on the solid content. To determine the cytotoxicity of AFWE on pulmonary alveoli, WST8 was performed on 3D4/31-PAM and A549-AEC. Although AFWE was not cytotoxic to 3D4/31-PAMs, A549-AECs viability was increased by 60 μg/mL AFWE and decreased by 6 μg/mL AFWE treatment (Fig. 1C). As the number of tissue-resident AMs decreases with the severity of lung infection [22,23], we tested the protective effect of AFWE on the proliferation of 3D4/31-PAMs. However, 60 μg/mL AFWE did not upregulate the proliferation of PMA-stimulated 3D4/31-PAMs (PS-3D4/31-PAMs) (Fig. 1D).
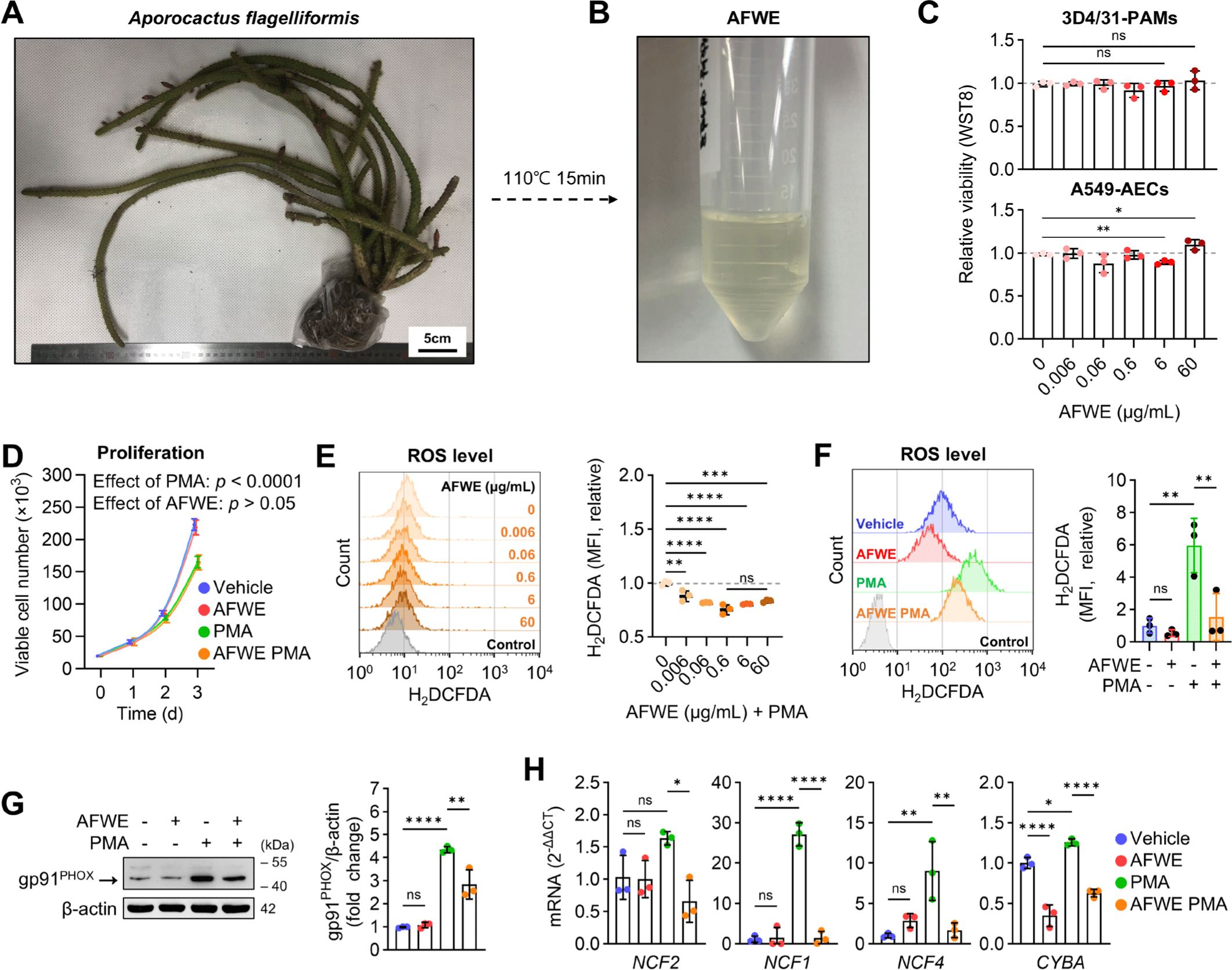
AFWE (0.006–60 μg/mL) reduced the intracellular ROS levels in PS-3D4/31-PAMs (Fig. 1E). 60 μg/mL AFWE showed no antioxidant effect on unstimulated 3D4/31-PAMs (Fig. 1F). This antioxidant effect of AFWE on PS-3D4/31-PAM is supported by the inhibition of NADPH oxidase 2 (NOX2) complex expression, a source of ROS, in inflamed AMs [24]. AFWE (60 μg/mL) suppressed the expression of NOX2 complex, including gp91PHOX (NOX2) (Fig. 1G), neutrophil cytosolic factor2 (NCF2), NCF1, NCF4, and cytochrome b-245 α chain (CYBA) in PS-3D4/31-PAMs (Fig. 1H). Overall, 60 μg/mL AFWE lowered ROS production via the downregulation of NOX2 without toxicity in 3D4/31-PAM. Furthermore, AFWE treatment at concentrations lower than 60 μg/mL was toxic in A549-AEC, so we established 60 μg/mL as the optimal AFWE concentration for alveolar immunomodulation and conducted subsequent experiments.
Next, we confirmed that AFWE inhibited proinflammatory gene expression and bactericidal activity in 3D4/31-PAMs. AFWE decreased the levels of proinflammatory markers such as COX2 (Fig. 2A), prostaglandin-endoperoxide synthase 2 (PTGS2), TNF, and interleukin 1 β (IL1B) (Fig. 2B) in PS-3D4/31-PAMs while slightly reducing the expression of the anti-inflammatory marker ARG1 and resistin-like β (RETNLB) (Fig. 2B). Bactericidal assays (Fig. 2C) revealed that AFWE diminished the bactericidal activity against E. coli DH5α in 3D4/31-PAMs (Fig. 2D and 2E). Moreover, AFWE reduced the E. coli-induced autophagy (Fig. 2F). These results indicate that AFWE downregulates proinflammatory features, including ROS production, proinflammatory gene expression, and bactericidal activity, in 3D4/31-PAMs.
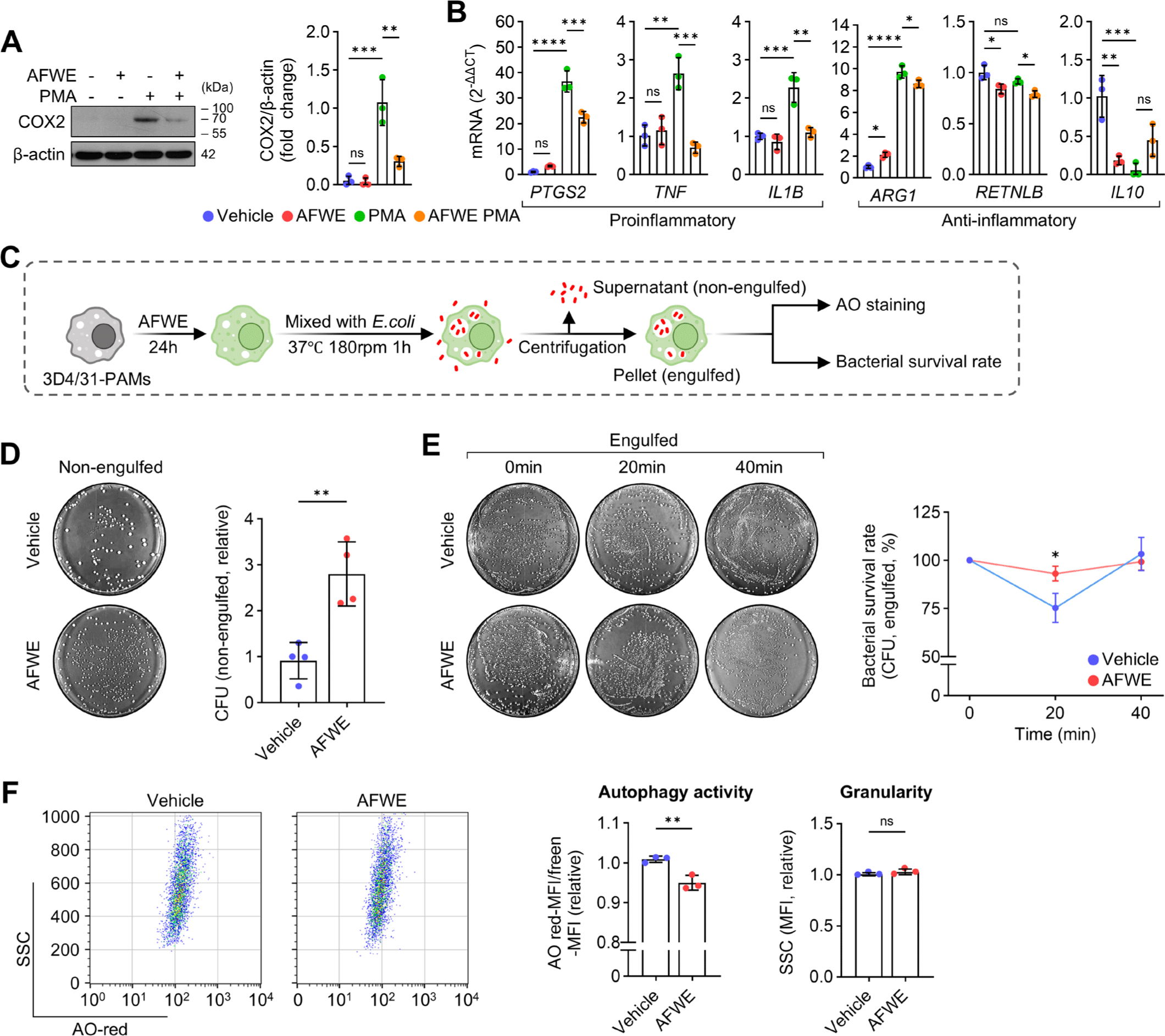
To confirm whether AFWE regulates P2Y14-associated metabolism in PAMs, glucose metabolism and P2Y14 cascade were quantified. Glucose uptake was reduced (Fig. 3A), and the LD content was upregulated (Fig. 3B and 3C) by AFWE in the 3D4/31-PAMs. The increased glycogen content in 3D4/31-PAMs following PMA stimulation was prevented by AFWE treatment (Fig. 3D and 3E). Considering that lipid accumulation in polarized macrophages depends on fatty acid uptake [25], our results indicate that AFWE selectively blocks P2Y14 (glycogen)-mediated inflammation. Glucose uptake, glycogenesis, and the pentose phosphate pathway (PPP) are essential for P2Y14-mediated proinflammatory responses. In particular, activation of the glycogenesis, characterized by intracellular glycogen accumulation, is required for the production of the P2Y14 ligand UDPG [10] (Fig. 3F). qRT-PCR showed that AFWE suppressed the expression of genes related to glucose uptake (SLC2A1, solute carrier family 2 member 1), glycogenesis (PGM1, phosphoglucomutase 1; PYGL, glycogen phosphorylase L; GAA, α-glucosidase), and PPP (G6PD, glucose-6-phosphate dehydrogenase; TKT, transketolase) in the 3D4/31-PAMs (Fig. 3G and 3H). AFWE suppressed P2RY14 and STAT1 expression in the PS-3D4/31-PAMs (Fig. 3I), suggesting that AFWE suppresses metabolism related to P2Y14 activation. These results suggest that AFWE suppresses P2Y14-associated proinflammatory features in PAMs.
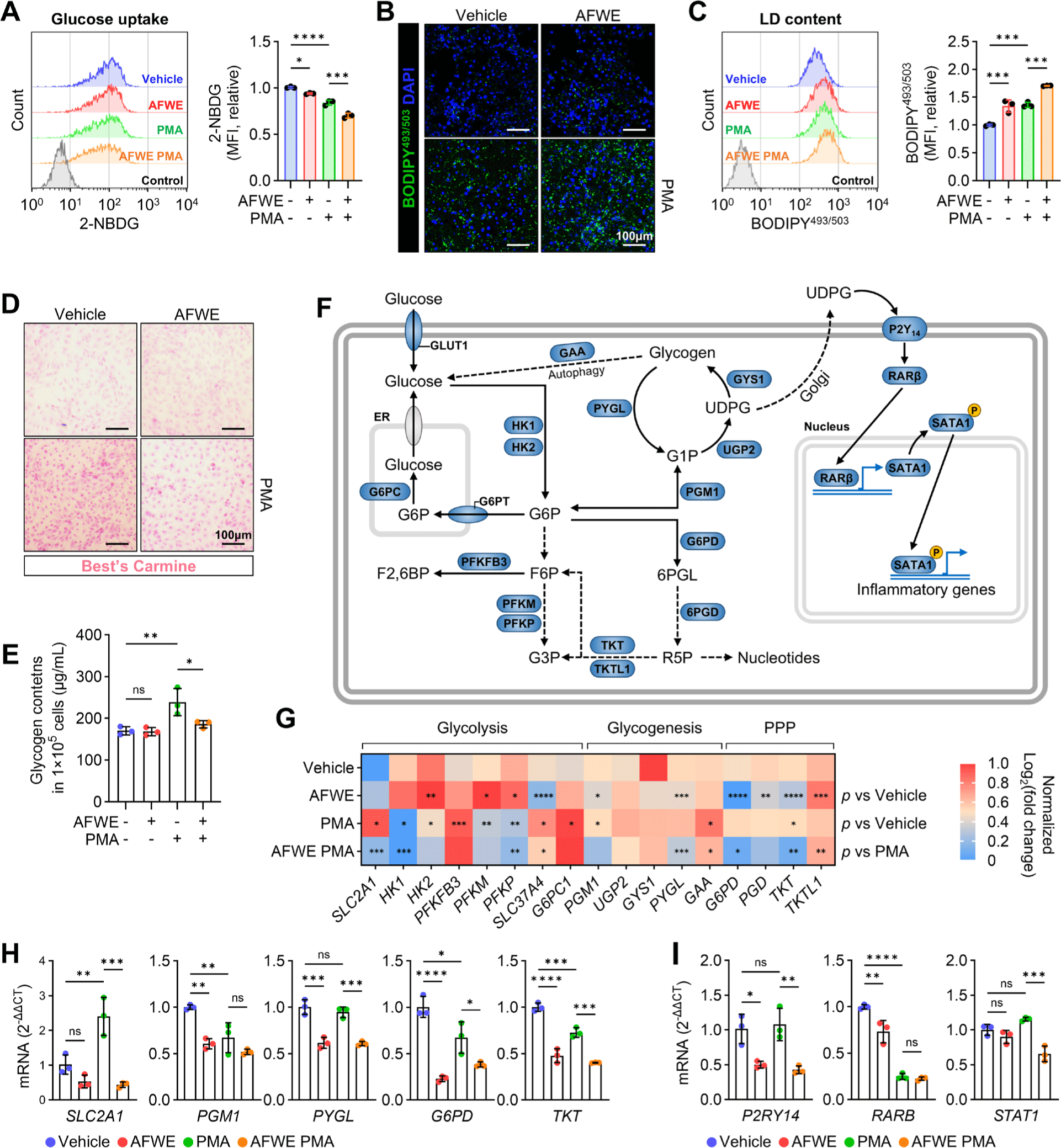
Polarity-based fractionation and LC-MS were performed to identify anti-inflammatory compounds in AFWE. The obtained fractions were concentrated 20-fold before use, and the dry weight of AFWE-OL3 was 52 mg/mL (Fig. 4A). AFWE-OL3 (52 μg/mL) reduced the levels of P2Y14 and PYGL in the immunoblotting of 3D4/31-PAMs (Fig. 4B). Genetic or chemical inhibition of PYGL can effectively inhibit P2Y14-mediated cytokine expression by reducing NADPH production [10]. The LC chromatogram of AFWE-OL3 showed a major peak at retention time (RT) 9.56 (Fig. 4C). The mass spectrum of RT9.56 was analyzed using the m/z CLOUD Mass Spectral Database, and the most similar compound was identified as limonin, also known as obaculactone and evodin (Fig. 4D). Based on the peak area (39%) at RT 9.56, AFWE-OL3 is expected to contain 28.42 mg/mL limonin. Considering that AFWE-OL3 was 20-fold enriched, it is estimated that limonin is present in AFWE at a concentration of 1.42 mg/mL. Limonin (Fig. 4E), a limonoid polyphenol found in citrus, has been reported to protect against lipopolysaccharide (LPS)-induced acute lung injury [26]. Interestingly, in citrus fruits, the glucose unit of UDPG can be transferred to limonin by limonoid glucosyltransferase [27].
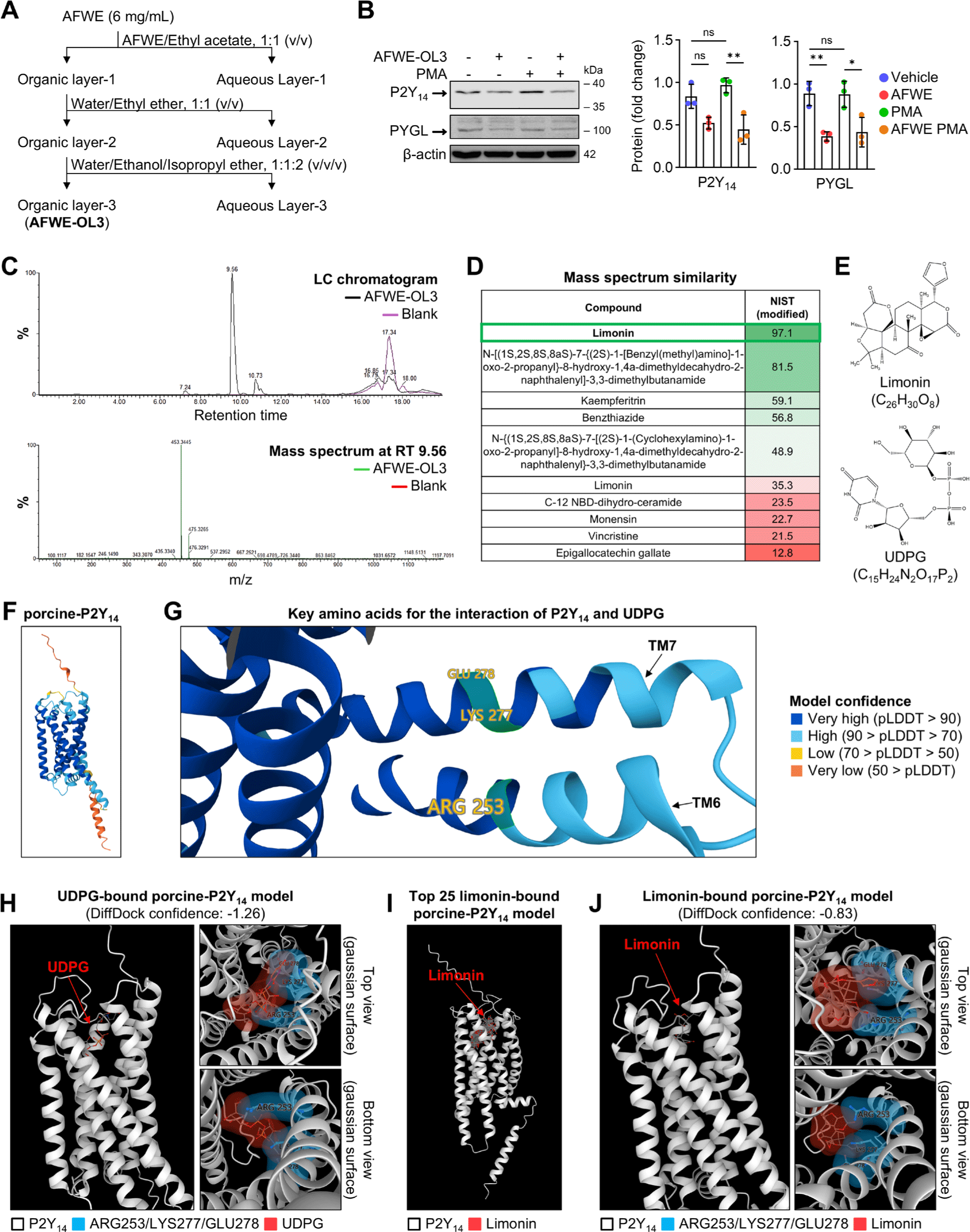
To assess the potential interaction between limonin and porcine-P2Y14, we performed computational molecular docking analysis. The structure of the porcine-P2Y14 (Fig. 4F) used in this study exhibited a very high confidence for ARG253/LYS277/GLU278 (Fig. 4G), a key amino acid in the interaction between P2Y14 and UDPG [28]. Our prediction showed that UDPG interacts with ARG253/LYS277/GLU278 in porcine-P2Y14 (Fig. 4H). The prediction of limonin docking to porcine-P2Y14 showed consistency in limonin poses (Fig. 4I). The prediction model with the highest score showed an interaction between limonin and ARG253/LYS277 (Fig. 4J). These results suggest the potential binding of limonin to the UDPG-binding site of porcine-P2Y14.
To confirm the anti-inflammatory effect of limonin on PAMs, we assessed the dose-response effect of limonin on viability and proinflammatory gene expression. Limonin treatment, at a final concentration of 30 μM, increased the viability of 3D4/31-PAMs cultured with or without PMA (Fig. 5A). Dose-response screening of limonin using qRT-PCR showed that 30 μM limonin suppressed the expression of P2RY14, STAT1, and PTGS2, but not ARG1, in PS-3D4/31-PAMs (Fig. 5B). Based on these results, we suggest that limonin at a final concentration of 30 μM has the potential to suppress P2Y14-mediated inflammation in PAMs.
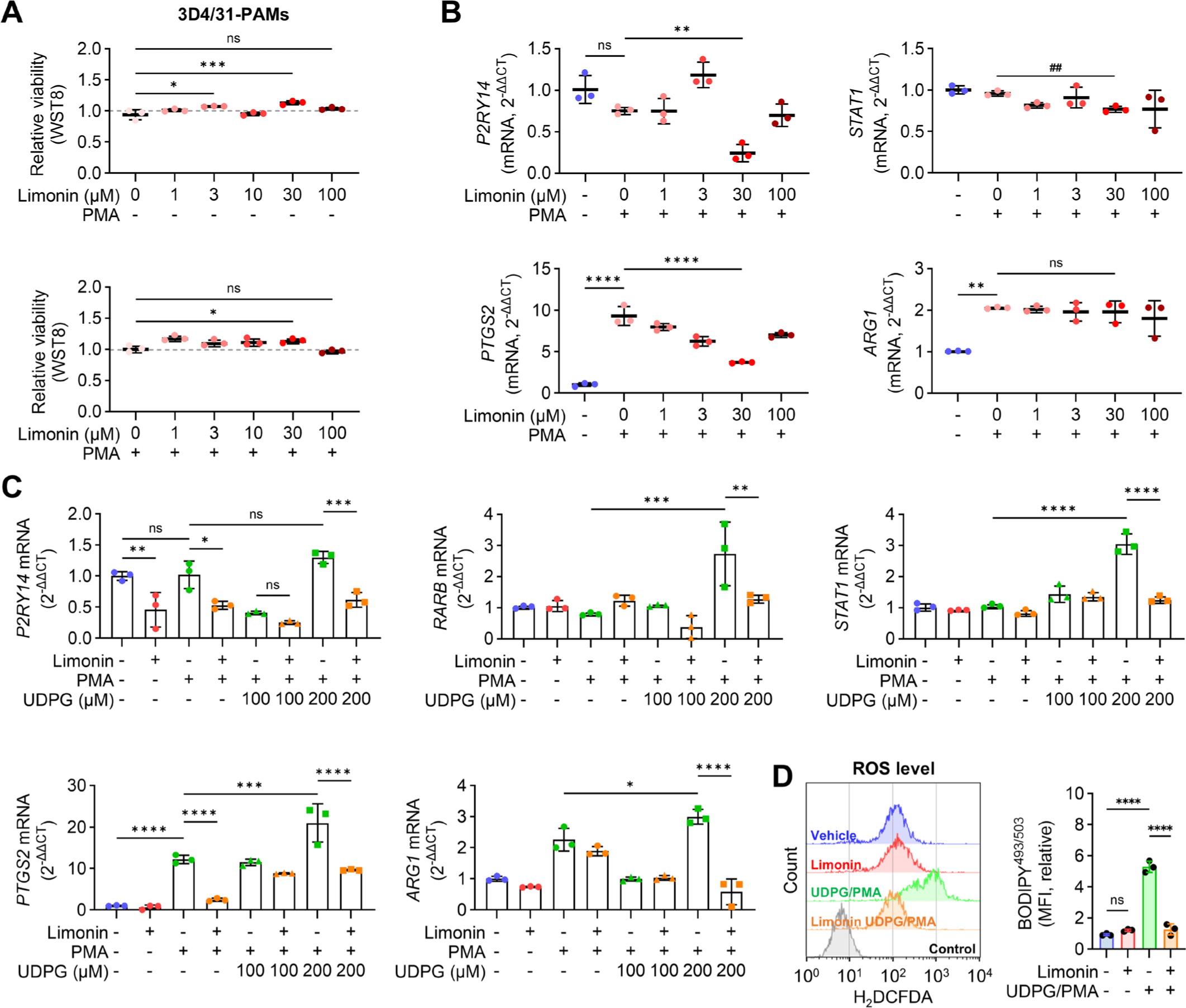
Next, to assesses the effects of limonin on UDPG-induced inflammation, 3D4/31-PAMs were stimulated with combination of PMA with UDPG. Significantly increased expression levels of retinoic acid receptor β (RARB), STAT1, PTGS2, and arginase-1 (ARG1) were observed in 3D4/31-PAMs stimulated with 200 µM UDPG. Limonin treatment suppressed the expression of P2RY14, RARB, STAT1, PTGS2, and ARG1 in 3D4/31-PAMs stimulated with PMA/UDPG (200 µM) (Fig. 5C). The PMA/UDPG-induced ROS production in 3D4/31-PAMs was reduced by limonin treatment (Fig. 5D). These results suggest that limonin has the potential to suppress UDPG/P2Y14-induced inflammation in PAMs.
DISCUSSION
PRDC remain the most serious threat to pig health and productivity. This study sought to explore the association of P2Y14 with porcine respiratory inflammation and to develop a new AID. AFWE inhibited ROS production by reducing the expression of NOX family members in PS-3D4/31-PAMs and suppressed glucose uptake and glycogenesis. We also demonstrated the potential of AFWE to inhibit P2Y14-mediated inflammation by reducing the expression of P2RY14, STAT1, TNF, ILB1, and PTGS2. Limonin reduced the UDPG-induced expression of P2RY14, RARB, STAT1, and PTGS2 in 3D4/31-PAMs. These results suggest the involvement of P2Y14 as a major regulator of inflammatory responses in PAMs and propose AFWE and limonin as AID candidates that can control this receptor.
Macrophages under inflammatory stimuli or after phagocytosis of bacteria increase cytokine and ROS production to recruit immune cells and eliminate pathogens [29]. AFWE inhibited NOX2 complex expression, ROS production, and bactericidal activity in 3D4/31-PAMs. These results are consistent with reports that NOX2 is a major source of ROS that kills phagocytic bacteria and that NOX2 deficiency impairs bactericidal activity [30]. AFWE inhibited the expression of proinflammatory genes PTGS2, TNF, and IL1B in 3D4/31-PAMs. TNF and IL1B induce macrophage activation and PTGS2 expression. COX2 (encoded by PTGS2) is a major target for anti-inflammatory drug development, as it plays a central role in the regulation of inflammatory processes through the modulation of vascular permeability and tissue swelling [31]. Therefore, the inhibitory effect of AFWE and limonin on PTGS2 expression suggests their potential as anti-inflammatory drugs. ARG1 expression was increased by PMA in 3D4/31-PAMs and slightly reduced by AFWE. ARG1 is classically used as a marker of anti-inflammation; however, AMs have been reported to express both inflammatory and anti-inflammatory markers and express high levels of ARG1 under chronic infection [32,33]. Therefore, we suggest that the increase in ARG1 levels by PMA and UDPG is due to the metabolic features of AMs.
Glucose is essential for energy metabolism and P2Y14-mediated inflammation. AFWE slightly decreased the glucose uptake of 3D4/31-PAMs but increased the LD content and had no effect on viability and proliferation. Given the high dependence of AMs proliferation and development on fatty acid metabolism [34], we suggest that activation of fatty acid metabolism may have maintained energy metabolism. PMA treatment increased SLC2A1 expression but decreased glucose uptake and did not increase P2Y14 cascade gene expression. PMA/UDPG treatment increased RARB and STAT1 expression, suggesting that P2Y14-mediated inflammation was activated. These results suggest that PMA is inadequate to induce P2Y14 activation at the mRNA expression level, and that PMA/UDPG combination treatment is suitable for P2Y14 activation in 3D4/31-PAM.
Increased P2Y14 activity is closely related to the exacerbation of various diseases including asthma [35], coronavirus disease 2019 [36], gouty arthritis [37], and intestinal inflammation [38], suggesting a variety of therapeutic uses for P2Y14 antagonists. In porcine, P2Y14 has been reported as a therapeutic target for heart disease and diabetes [11,18], and we demonstrated its role in porcine alveolar immunity. We observed increased glycogenesis, a characteristic feature of P2Y14-mediated inflammation, in PS-3D4/31-PAMs. These results suggest an increased glucose requirement by macrophages in inflammatory responses, and are consistent with LPS-induced increased glucose consumption and hypoglycemia [39]. Additionally, porcine skeletal muscle growth rate is associated with the expression of glycogenesis-related genes (PGM1, phosphoglucomutase 1; UGP2, UDPG pyrophosphorylase2) [40]. These findings make it interesting to study the effects of P2Y14 and UDPG levels on porcine productivity.
The selection of extraction solvents considers various factors such as extraction efficiency, environmental hazards, and residual toxicity. In line with the global trend toward eco-friendly industries, the importance of water extraction technology is increasing [41]. Succulent Opuntia species, known to have potential as livestock feed [13], have been reported to lack antioxidant and antibacterial effects in aqueous extracts [42]. This is attributed to the low solubility of major bioactive compounds such as polyphenols in water [43]. Using LC-MS, we identified limonin as a potential bioactive compound in AFWE. Limonin can be extracted from Citrus grandis (pomelo) via water and resin absorption [44]. Additionally, limonin is abundant in the peel of Citrus aurantifolia (lime), which is often discarded as waste and can be extracted using the low-toxicity solvent, ethanol [45]. In this study, the limonin content of AFWE estimated by LC-MS was 23.69% (w/v), 1.42 mg/mL. A 14.21 μg/mL of limonin content was expected with 60 μg/mL AFWE treatment, which was similar to the optimal anti-inflammatory activity concentration of 30 μM (14.12 μg/mL) of limonin. This suggests that limonin confers anti-inflammatory activity to AFWE. Overall, these findings suggest that AFWE and limonin are promising AID candidates for the eco-friendly livestock industry.
Limonin has been noted for its various pharmacological activities but has limited clinical potential due to unclear mechanisms of action [46]. In this study, we confirmed that AFWE and limonin reduced PMA-induced expression of PTGS2, STAT1, and P2RY14 in 3D4/31-PAMs. Our molecular docking prediction also indicated that limonin has a higher binding score to porcine-P2Y14 than to UDPG. These results suggest that the potential mechanism of action (MOA) of AFWE and limonin involves binding to and inhibiting the activity of porcine-P2Y14. Further research is required to clarify this MOA, including confirmation of the nuclear localization of STAT1/RARβ, which is characteristic of P2Y14 activation [10]. Our molecular docking analysis used the predicted porcine-P2Y14 structure due to the limited number of studies on porcine-P2Y14. Although UDPG binding to P2Y14 induces structural changes [28], mechanical binding studies of limonin to porcine-P2Y14 have not been conducted. Nevertheless, the reduction in the UDPG-induced expression of RARB, STAT1, and PTGS2 in 3D4/31-PAMs by limonin suggests the potential of limonin to inhibit P2Y14-mediated inflammation.
In summary, AFWE exerted anti-inflammatory effects in 3D4/31-PAMs, inhibiting ROS production and NOX2/PTGS2/TNFA expression reported in PRDC. Limonin, a compound identified from AFWE, inhibited the UDPG-induced expression of P2Y14 cascade genes and PTGS2 in 3D4/31-PAMs. Overall, our results suggest that P2Y14 is a target for the control of PRDC and provides new insights into the inflammatory response of PAMs. Furthermore, we propose AFWE and limonin as candidate AIDs for porcine.
















