INTRODUCTION
Porcine epidemic diarrhea virus (PEDV) is a highly virulent pathogen that poses a significant threat to the swine industry, particularly to neonatal piglets [1–3]. PEDV infection causes severe gastrointestinal symptoms, including diarrhea, vomiting, and dehydration, which can lead to mortality rates approaching 100% in affected populations [4,5]. The virus primarily targets the intestinal epithelium [6], particularly in the jejunum and ileum, leading to villous atrophy and compromised gut integrity [7]. Understanding PEDV pathogenesis is essential for developing effective therapeutic strategies and vaccines.
Traditionally, PEDV research has relied on two-dimensional (2D) cell-culture models, such as the IPEC-J2 and IPI-2I cell lines derived from porcine intestinal tissue [8,9]. However, those models are limited in their ability to replicate the complex in vivo environment of the porcine intestine, which features diverse cell types, unique structural architecture, and region-specific physiological characteristics [10]. Recently, organoid technology has presented a promising alternative, allowing for the cultivation of three-dimensional (3D) organoids that more accurately mimic the physiological conditions of the intestinal epithelium [4,11,12]. Importantly, organoids can be derived from specific regions of the intestine, such as the duodenum, jejunum, and ileum, enabling the study of region-specific responses to PEDV infection [13].
Studies have shown that PEDV infection efficiency and host responses vary across intestinal segments, likely due to inherent differences among the duodenum, jejunum, and ileum [14,15]. Furthermore, distinct gene expression patterns in the small intestine suggest that each region has specialized physiological functions that might influence its susceptibility to infection [16]. Despite the increasing use of organoids to study enteric viruses, few studies have investigated the region-specific gene expression responses to PEDV infection, and the influence of those differences on viral pathogenesis remains underexplored [17–19].
For this study, we established apical-out porcine intestinal organoids derived from the duodenum, jejunum, and ileum to investigate region-specific transcriptional and functional responses to PEDV infection. Specifically, we investigated how different intestinal regions responded to PEDV at the molecular level, identifying variations in gene expression profiles, immune activation, and antiviral defense mechanisms.
MATERIALS AND METHODS
All procedures involving pigs were approved by the Seoul National University Institutional Animal Care and Use Committee (SNU-230915-4-1) and conducted in accordance with guidelines for the care and use of laboratory animals. The PEDV DR13 strain used in this study was generously provided by Professor Daesub Song from Seoul National University [20].
Vero cells were cultured in Dulbecco modified Eagle medium (DMEM, Biowest) supplemented with 5% fetal bovine serum (FBS, Biowest) and 1% antibiotic-antimycotic (Gibco) in a 37°C incubator supplied with 5% CO2. Vero cells were subcultured at 70%–80% confluency, and PEDV was inoculated when the cells reached 70%–80% confluency.
The virus was propagated in Vero cells that were seeded one day before infection in a T-75 flask at 70%–80% confluency and incubated overnight at 37°C with 5% CO2. The cells were washed once with phosphate-buffered saline (PBS, Biosesang) and inoculated with diluted PEDV DR13 for 1 h. Then the cells were washed once with PBS and maintained in the PEDV infection medium, which contained DMEM (Biowest) with 0.3% tryptose phosphate broth (Becton Dickinson), 0.02% yeast extract (Gibco), 2 µg/mL TPCK trypsin (Thermo Fisher Scientific), and 1% antibiotic-antimycotic (Gibco) at 37°C in 5% CO2. The cytopathic effects (CPE) were monitored daily, and the virus was harvested using three rounds of freezing and thawing after 72–96 h, when the CPE exceeded 80%. The cell supernatant was centrifuged at 4,000 rpm for 20 min, and then the supernatant of the sample was aliquoted and stored at –80°C until use. The virus titer was measured using Vero cells that were seeded in 96-well plates at a density of 2 × 104 cells/well in 200 µL of culture medium and incubated at 37°C in 5% CO2. The virus was diluted in a 10-fold series in DMEM with 1% antibiotic-antimycotic. Each dilution was added to the well in six replicates, and the negative control cells were treated only with DMEM and 1% antibiotic-antimycotic. After 1 h of incubation with the virus, the cells were washed once with PBS and then maintained in the PEDV infection medium, as in the propagation procedure. After 72–96 h, when the CPE exceeded 80%, the mean tissue culture infection dose 50 (TCID50) was calculated using the Spearman-Kärber method.
Duodenum, jejunum, and ileum tissue from of small intestines of 1-week-old piglets was dissected, and crypts were isolated with Gentle cell dissociation reagent (StemCell Technologies). Then, the crypts were embedded in Matrigel (Corning) with growth medium. The organoids were passaged every 4–5 days. To differentiate the porcine intestinal organoids, the culture medium was changed to differentiation medium after 2–3 days of passaging.
The porcine intestinal organoid growth medium was Advanced DMEM (Gibco) supplemented with 2 mM GlutaMAX, 10 mM HEPES (Gibco), 1% penicillin/streptomycin, 1X N-2 supplement, 1X B-27 supplement without vitamin A (Thermo Fisher Scientific), 500 ng/mL human R-spondin 1, 100 ng/mL human Noggin, 50 ng/mL human EGF (PeproTech), 100 ng/mL WNT surrogate-Fc fusion protein (ImmunoPrecise Antibodies), 0.5 µM A83-01, 10 µM SB202190, 10 mM nicotinamide, 10 nM human gastrin I, 5 µM LY2157299, 2.5 µM CHIR99021 (Sigma-Aldrich), and 10 µM Y-27632 (MedChemExpress).
The porcine intestinal organoid differentiation medium contained reduced concentrations of human R-spondin 1, human Noggin, and WNT surrogate-Fc fusion protein (1/10 in the culture medium) and 10 mM DAPT (Sigma-Aldrich).
The organoids were passaged 2–3 days prior to starting the apical-out culture and were maintained in growth medium. Matrigel-embedded organoids have basolateral surfaces facing outward, which are referred to as basal-out organoids. To generate apical-out organoids, the organoids were first separated from the Matrigel by incubating them in 5 mM EDTA in PBS on a shaking rotor for 30 min at 4°C to remove the extracellular matrix proteins. The organoids were centrifuged at 200×g for 5 min at 4°C. Then, the pellet was re-suspended in differentiation medium in ultra-low attachment 24-well cell culture plates (Corning). After transitioning to apical-out culture for 3 days, the suspended organoids exhibit reversed polarity, with the apical surfaces face outward. These are referred to as apical-out organoids.
We infected 200 apical-out organoids with PEDV DR13 for 1 h at 37°C. After 1 h, the medium containing the virus was removed, and differentiation medium was added.
The basal-out organoids were separated from the Matrigel and infected with PEDV DR13 in ultra-low attachment 24-well plates for 1 h at 37°C. After 1 h, these organoids were re-embedded in Matrigel, and differentiation medium was added. Sampling was conducted 24 h after inoculation.
The cultured apical-out organoids were harvested, and total RNA was extracted using Trizol reagent (Invitrogen). From each sample, 2 µg of RNA was taken and converted into cDNA using M-MLV reverse transcriptase (Promega). Real-time quantitative PCR was conducted as previously described [21] on a CFX Duet (Bio-Rad) with SYBR green master mix (Applied Biosystems). Porcine GAPDH was used to normalize gene expression. The sequences of the primers used in this study are listed in Table 1.
Organoids separated from the Matrigel were fixed in 4% paraformaldehyde for 20 min. For cryo-sectioning, the fixed organoids were embedded first in 30% sucrose and subsequently in a gelatin/sucrose solution. Immunocytochemistry staining was conducted on 12-μm sections of the gelatin-embedded organoids. These sections were incubated in blocking/permeabilization buffer (1.5 mL of FBS, 0.5 g of bovine serum albumin (BSA, LPS solution) 250 μL of Triton X-100, 250 μL of Tween 20, and 500 μL of 1% (wt/vol) sodium deoxycholate solution in 47.5 mL of PBS [22]) for 1 h at room temperature. Primary antibodies were then applied overnight at 4°C. After the samples were washed, secondary antibodies were applied for 2 h at room temperature. Then, the samples were counterstained with DAPI (Sigma-Aldrich) and mounted with fluorescence mounting medium (Dako). Fluorescent images were obtained using a Leica TCS SP8 X (Leica Microsystems). The antibodies used in this study are listed in Table 2.
Sequencing of the extracted RNA was performed at Macrogen Incorporated using the manufacturer’s reagents and protocol. mRNA sequencing libraries were prepared from the extracted RNA by using an Illumina TruSeq stranded mRNA sample prep kit (Illumina). Indexed libraries of the samples were submitted to paired-end read sequencing on an Illumina NovaSeqX (Illumina). The sequenced data were processed and analyzed with minor modifications to a previously described procedure [23]. Adapter sequences were removed and low-quality reads were filtered using Cutadapt (v4.9). The trimmed sequences were aligned to the Sus scrofa reference genome (susScr2) using HISAT2 (v2.2.1) and counted by featureCounts (v2.0.3). Gene expression was quantified by EdgeR (v3.36.0), and differentially expressed genes (DEGs) were further analyzed. We identified DEGs using absolute Log2 fold change≥1 and p-values<0.05 as the threshold. For the Gene Ontology (GO) analysis of DEGs, PANTHER19.0 [24] was used to categorize the genes into Panther GO terms. The DEGs were also annotated into KEGG database pathways using ShinyGO 0.81 (http://bioinformatics.sdstate.edu/go/) [25,26]. Volcano plots of the DEGs were generated with ggplot 2 in R. A principal component analysis (PCA) plot and heatmaps were generated using an in-house script in R. For the heatmaps, the samples were clustered according to Euclidean distance. Proportional Venn diagrams of up- and downregulated genes were created using BioVenn [27]. The RNA-seq data presented in this study have been deposited in the NCBI GEO database under accession number GSE 280630.
All experiments were performed in at least triplicate. Depending on the number of groups to be compared, t-tests and one-way ANOVA were used to analyze the data. The values are represented as the mean ± SEM. A p-value of less than 0.05 were considered statistically significant. All statistical analyses were performed using GraphPad Prism software (v8.0.1).
RESULTS
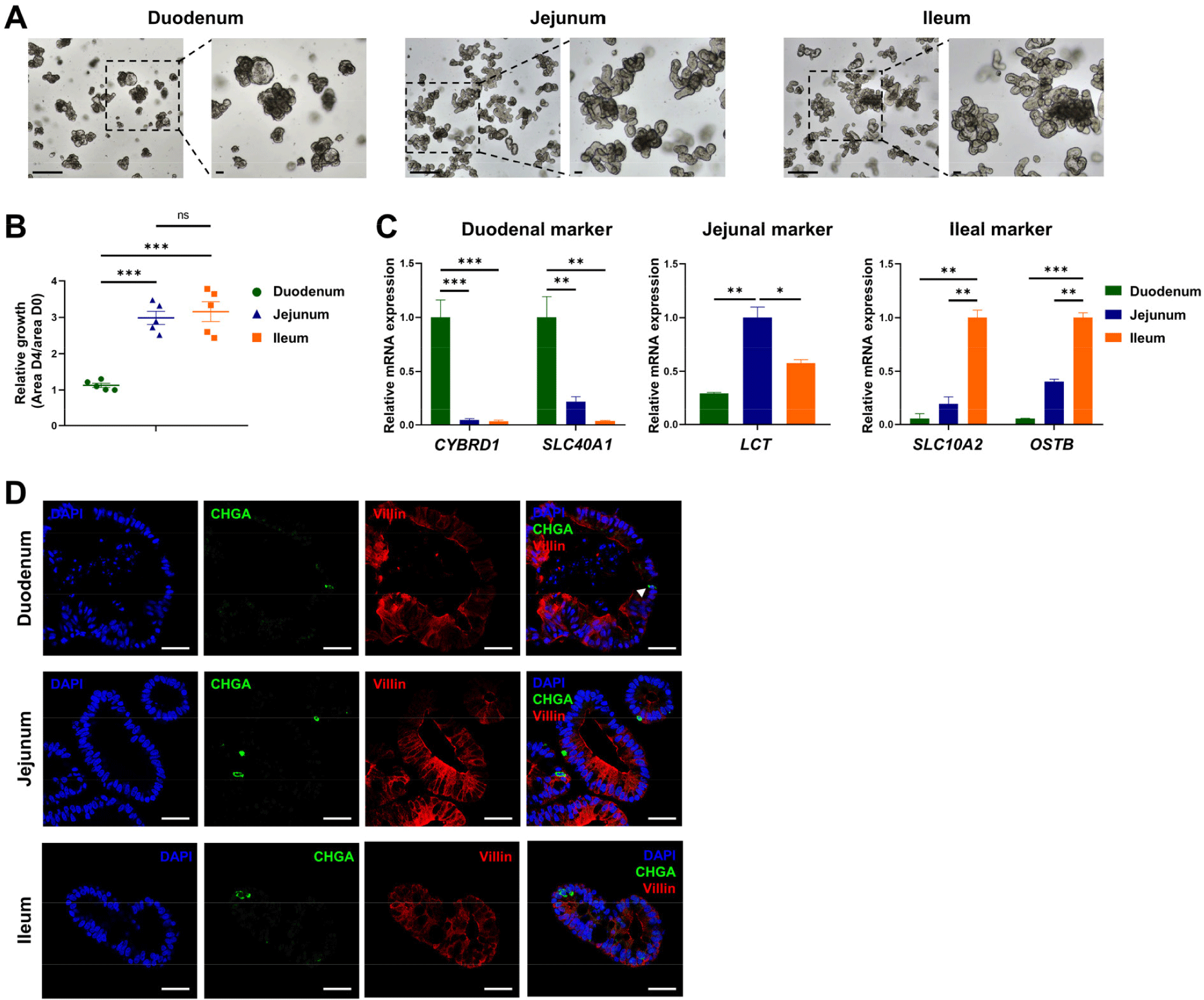
To develop an in vitro model of the porcine intestine, we established organoids derived from the duodenum, jejunum, and ileum. A microscopy morphological analysis revealed distinct structural features in the organoids from each region, with duodenal organoids resembling the structure of gastric organoids and showing morphological differences from the jejunal and ileal organoids (Fig. 1A). The proliferation analysis showed significantly lower proliferation rates in the duodenal organoids than in the jejunal and ileal organoids (Fig. 1B). Consistent with previous findings, these structural and functional differences across regions could contribute to differential responses to PEDV infection [14,15]. The qRT-PCR analysis of region-specific markers confirmed the establishment of region-specific organoids: the duodenal organoids expressed high levels of CYBRD1 and SLC40A1, jejunal organoids showed elevated levels of LCT, and ileal organoids had increased expression of SLC10A2 and OSTB (Fig. 1C). Immunofluorescence staining revealed consistent expression of the enterocyte marker Villin and the enteroendocrine marker chromogranin A (CHGA) across all three region-derived organoids (Fig. 1D). Notably, enterocytes, the primary target of PEDV, were present in all organoids, indicating that the developed organoids closely replicate the in vivo porcine intestine environment.
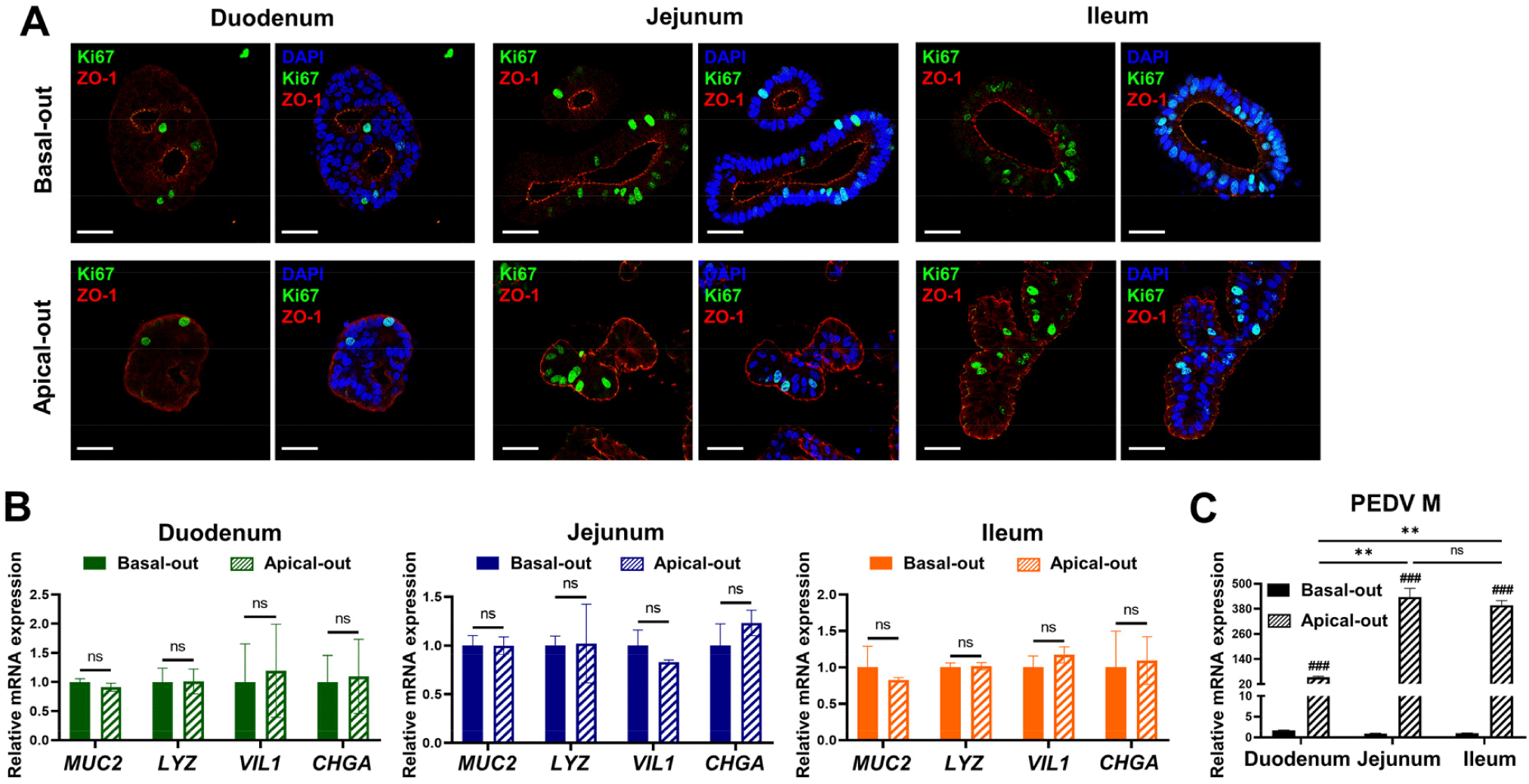
To investigate PEDV replication and host responses, we cultured apical-out and basal-out organoids derived from the duodenum, jejunum, and ileum. Immunofluorescence staining was performed to assess polarity, and it showed the apical marker ZO-1 on the outer membrane in the apical-out organoids and the inner membrane in the basal-out organoids, confirming the correct polarity. Additionally, the proliferation marker Ki67 was more prominent in the jejunal and ileal organoids, aligning with the proliferation patterns observed in Fig. 1B (Fig. 2A). To characterize the cell composition, we analyzed the expression of cell-specific markers by qRT-PCR and found no significant differences between the basal-out and apical-out organoids. Goblet cell (MUC2), Paneth cell (LYZ), enterocyte (VIL1), and enteroendocrine cell (CHGA) markers were all detected, indicating the presence of diverse cell types across organoids of both polarities in each intestinal region (Fig 2B). The PEDV genomic RNA analysis showed more robust PEDV replication in the apical-out organoids, particularly the jejunal and ileal organoids, than in the basal-out and duodenal organoids (Fig. 2C). To assess host responses to PEDV infection, the expression levels of inflammation-related markers (CXCL9, CXCL10, and IL17) and interferon pathway genes (ISG15, ISG58, and IFNL3) were measured. These markers were less expressed in the duodenal organoids than in the jejunal and ileal organoids (Supplementary Fig. S1A), suggesting that the jejunal and ileal organoids were more responsive to PEDV infection.
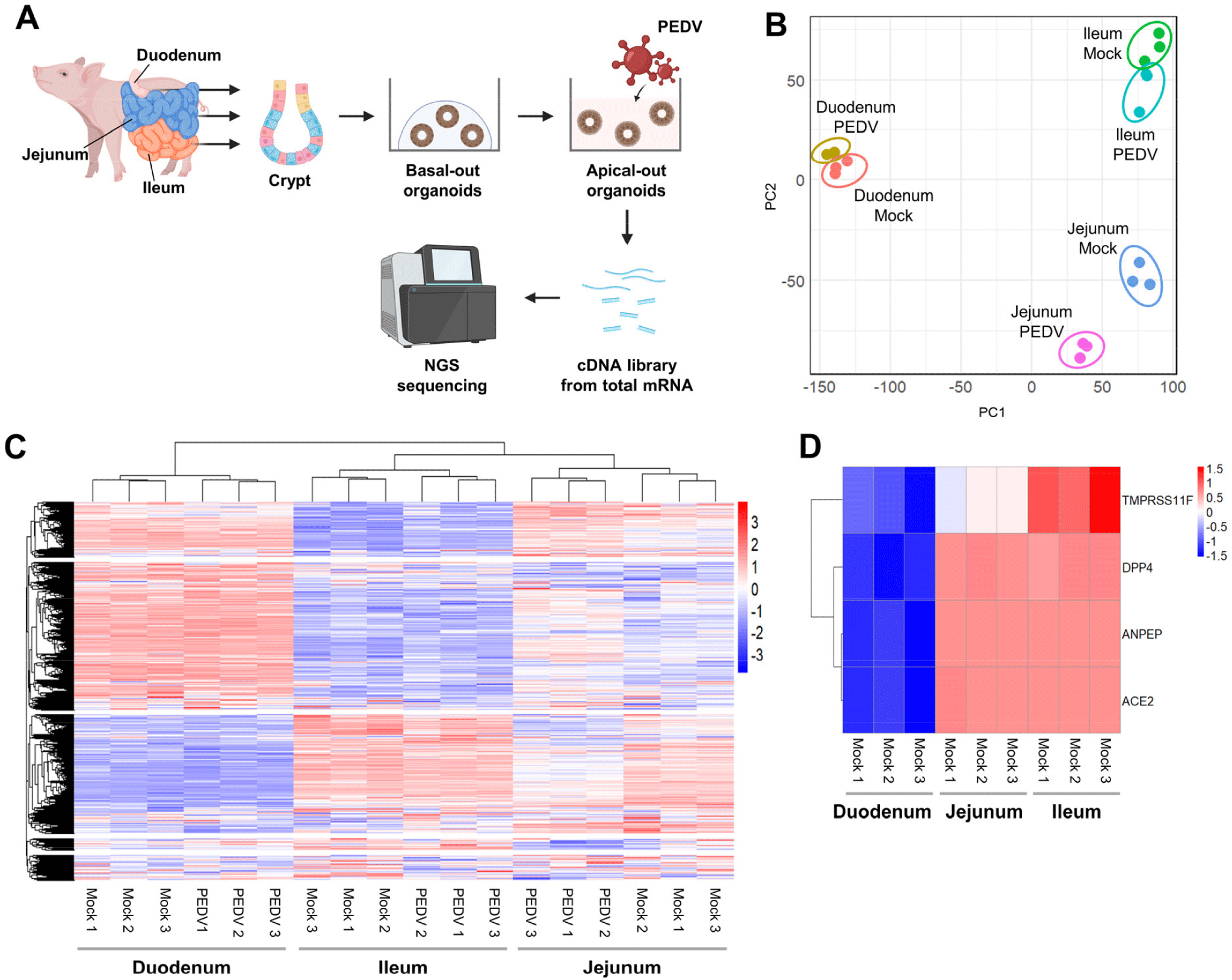
To further investigate regional responses to PEDV at the transcriptomic level, we performed RNA sequencing on apical-out organoids from each region (Fig. 3A). The PCA demonstrated distinct transcriptomic profiles by region and infection status, indicating unique regional responses to PEDV infection (Fig. 3B and Supplementary Fig. S2A, S2B, and S2C). Heatmap clustering indicated close gene expression profiles between jejunal and ileal organoids, with duodenal organoids forming a separate cluster, even in the absence of infection (Fig. 3C). Notably, key receptors and entry-related genes (ANPEP, ACE2, and DPP4) were more highly expressed in jejunal and ileal organoids than in duodenal organoids in mock conditions, as were the proteases TMPRSS4 and TMPRSS11F, which facilitate viral entry (Fig.3D). These findings suggest region-specific baseline gene expression profiles and receptor availability that might influence PEDV susceptibility.
To investigate region-specific transcriptional responses to PEDV infection, we analyzed DEGs in the duodenal, jejunal, and ileal organoids. A total of 20,428 genes were profiled, and DEGs with significant changes between the PEDV-infected and mock groups were identified using a threshold of p<0.05 and |Log2 fold change| ≥1. A volcano plot shows significant upregulation and downregulation of genes across regions (Fig. 4A). Specifically, the duodenum exhibited 58 upregulated and 40 downregulated genes, the jejunum had 101 upregulated and 261 downregulated genes, and the ileum had 67 upregulated and 131 downregulated genes (Fig. 4B). The GO enrichment analysis revealed region-specific functional categories among the DEGs. The jejunal and ileal organoids showed enrichment in GO terms related to cellular processes, biological regulation, metabolic activity, and membrane components, and the duodenal organoids had fewer gene counts in those enriched terms (Fig. 4C).
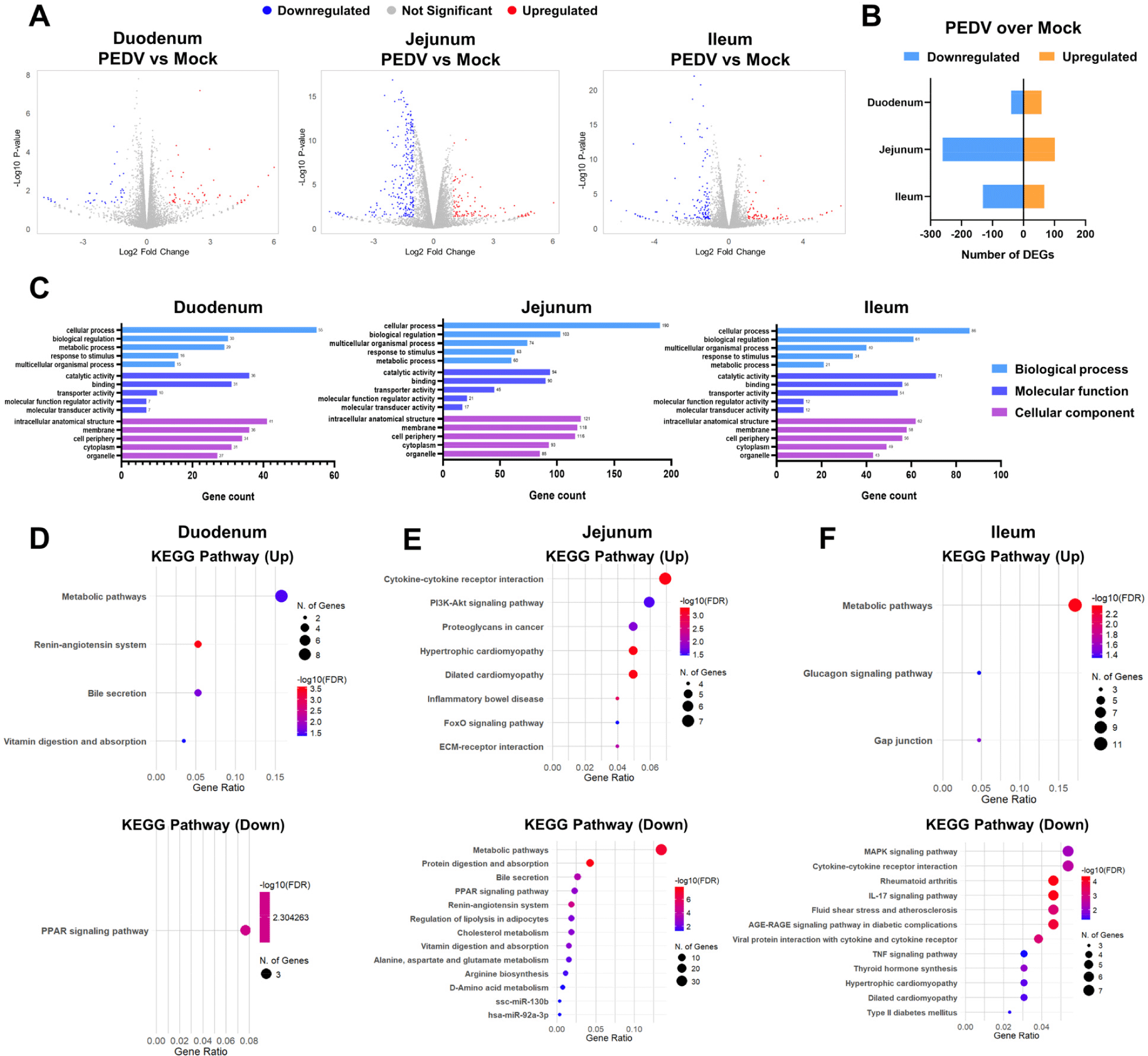
The KEGG pathway analysis further highlighted these region-specific responses. In the duodenum, the upregulated genes were associated with metabolic pathways and the renin-angiotensin system, and the downregulated genes were linked to PPAR signaling (Fig. 4D). In the jejunum, the upregulated genes were enriched in cytokine–cytokine receptor interactions and PI3K-Akt signaling, and the downregulated genes were involved in metabolic pathways and protein digestion and absorption (Fig. 4E). In the ileum, the upregulated genes were associated with metabolic pathways and the glucagon signaling pathway, and the downregulated genes were linked to the MAPK signaling pathway and cytokine–cytokine receptor interactions (Fig. 4F). These findings demonstrate distinct transcriptional responses to PEDV infection across intestinal regions, with each segment activating unique pathways.
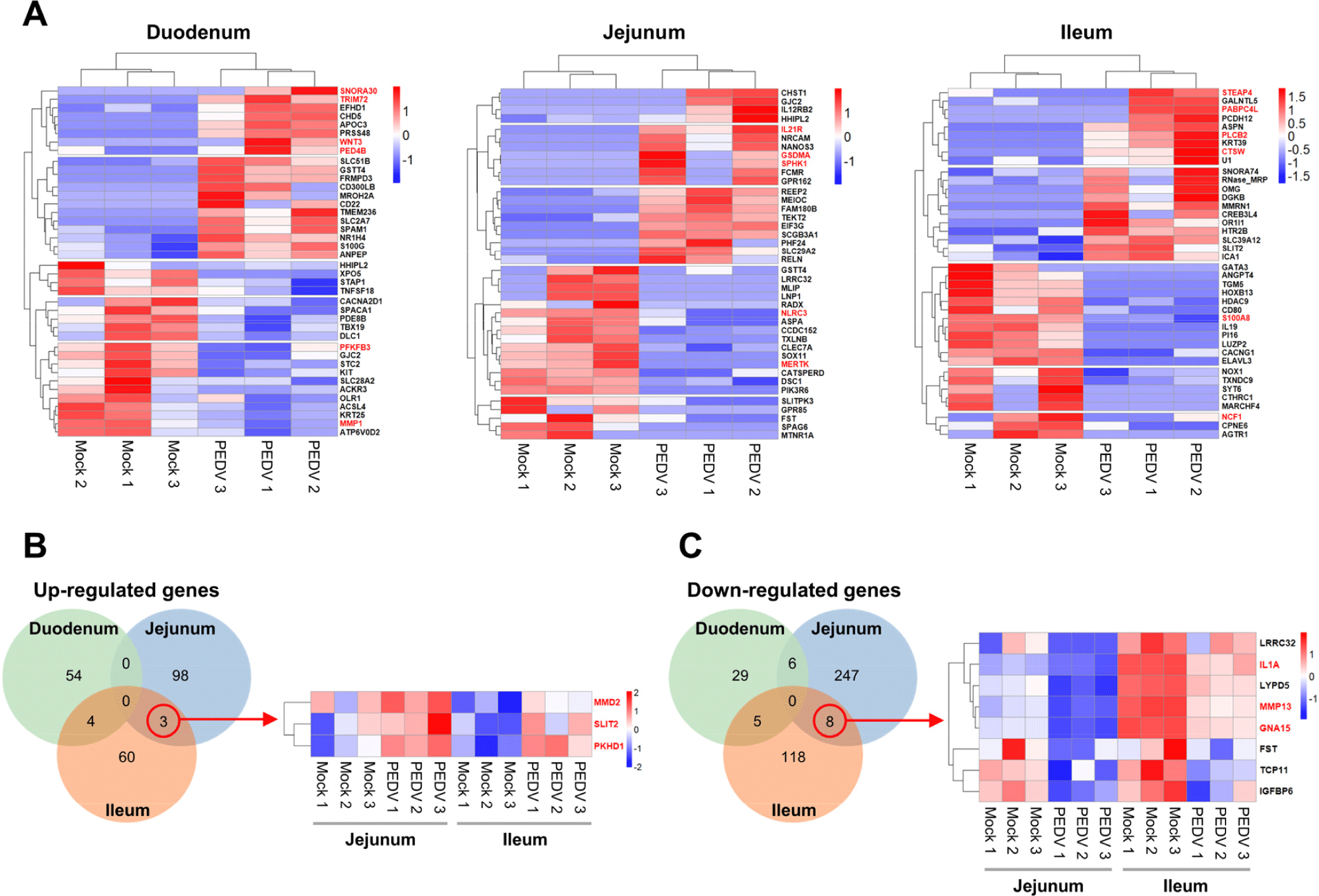
To further investigate the regional differences in gene expression responses to PEDV infection, we identified the top 20 genes upregulated and downregulated in the duodenal, jejunal, and ileal organoids, compared with the mock groups. A heatmap analysis revealed distinct expression patterns across regions (Fig. 5A). In PEDV-infected duodenal organoids, upregulated genes such as SNORA30 (viral processes), TRIM72 (viral restriction), WNT3 (Wnt/β-catenin signaling), and PDE4B (inflammatory response) suggest an immune response, and downregulated genes such as PFKFB3 (carbohydrate metabolism) and MMP1 (extracellular matrix remodeling) indicate reduced metabolic and cellular activity. In the jejunal organoids, genes such as IL12R (immune response), GSDMA (cell death), and SPHK1 (sphingolipid metabolism) were upregulated, reflecting active immune and inflammatory responses, and the downregulated genes, such as NLRC3 (PI3K-mTOR inhibition) and MERTK (viral entry), point to modulated immune signaling. In the ileal organoids, upregulated genes such as STEAP4 (antiviral response), PABPC4L (coronavirus inhibition), PLCβ2 (inflammation regulation), and CTSW (viral escape mechanisms) indicate robust immune activation, and the downregulated genes, such as NCF1 and S100A8 (inflammation modulation), show an adjusted inflammatory response. Venn diagrams of the DEGs reveal regional specificity, with three genes (SLIT2, MMD2, and PKHD1) commonly upregulated in the jejunal and ileal organoids (Fig. 5B). SLIT2 reduces inflammation and tissue damage, and PKHD1 supports epithelial barrier integrity, reflecting a coordinated response to maintain immune balance and tissue resilience in these regions. Among the 8 genes most commonly downregulated in the jejunal and ileal organoids (Fig. 5C), IL-1A, MMP13, and GNA15 are involved in immune regulation and viral response, indicating inflammation and an antiviral response to PEDV infection. These commonly altered genes likely play key roles in the jejunum and ileum, where PEDV infection is prominent.
DISCUSSION
This study provides critical insights into the region-specific responses of porcine intestinal organoids derived from the duodenum, jejunum, and ileum to PEDV infection. Using an apical-out organoid model, we observed distinct functional responses to PEDV across these intestinal regions, suggesting that the jejunum and ileum might be especially critical for PEDV pathogenesis in neonatal piglets. These findings indicate that regional susceptibility to PEDV is influenced not only by anatomical differences but also by the unique transcriptomic profiles of each segment.
Our quantitative analysis revealed significantly higher PEDV replication in jejunal and ileal organoids than in duodenal organoids, supporting our hypothesis that susceptibility is modulated by region-specific gene expression patterns. Bulk RNA sequencing confirmed a more robust activation of immune and antiviral pathways in the jejunum and ileum, implying an elevated capacity for viral response in these regions. This finding aligns with other studies that show differential immune responses across intestinal segments, which could inform more effective intervention strategies [14,28,29].
Recent studies indicate that aminopeptidase N (APN) alone does not fully account for PEDV entry because infection persists in APN-knockout cells and pigs [30–32]. Our findings corroborate that, showing that the jejunal and ileal organoids had significantly higher baseline expression of viral entry receptor genes, including ANPEP, ACE2, and DPP4, and suggesting that, beyond APN, other known coronavirus receptors such as DPP4 and ACE2 might contribute to heightened intestinal infection. These findings are consistent with previous studies showing that DPP4 and ANPEP expression is elevated in the jejunum compared with other regions [16,33]. Additionally, the increased expression of proteases such as TMPRSS4 and TMPRSS11F in these segments likely contributes to their increased susceptibility to PEDV, highlighting the importance of receptor availability in determining viral tropism [34,35].
In response to PEDV infection, we observed significant regulation of genes involved in immune modulation and tissue integrity. Notably, SLIT2, MMD2, and PKHD1 were upregulated in the jejunal and ileal organoids, suggesting a coordinated response to control inflammation and maintain tissue structure. SLIT2 has been recognized for its anti-inflammatory properties, suggesting that its upregulation could play a critical role in balancing immune activation with tissue preservation during PEDV infection [36]. The downregulation of pro-inflammatory genes such as IL-1A and MMP13 further indicates a protective mechanism that limits tissue damage [37–40]. This expression pattern supports a finely tuned immune response that promotes effective antiviral activity while preserving tissue integrity, positioning the jejunal and ileal regions as crucial sites of balanced, protective immune responses during PEDV infection.
This study highlights the utility of 3D organoid models in representing region-specific PEDV infection dynamics more accurately than traditional 2D models. While our focus was on porcine intestinal organoids, comparing our findings with human and other animal models offers broader insights. For example, receptors like ACE2 and DPP4 are not only involved in PEDV entry in pigs but also play a significant role in the entry of coronaviruses such as SARS-CoV-2 in humans. Similar upregulation patterns of these receptors have been observed in both species, indicating conserved mechanisms of viral entry. Understanding these cross-species similarities and differences may refine therapeutic strategies and broaden our comprehension of coronavirus-host interactions. Our findings suggest that therapeutic strategies might be optimized by tailoring them to target specific intestinal regions, particularly the jejunum and ileum, where PEDV susceptibility is high. However, our current organoid model lacks immune cells, which are essential for simulating in vivo immune responses. Interactions between the intestinal epithelium and immune cells such as T cells, macrophages, and dendritic cells are crucial for mediating host responses to viral infections [41,42]. The absence of these components may limit the model’s ability to fully replicate immune-mediated aspects of PEDV pathogenesis. Future studies could enhance these models by incorporating immune cells to better reflect in vivo conditions. Additionally, co-culturing with other tissue organoids may provide a more comprehensive system to investigate the systemic effects of PEDV across multiple tissues.
In conclusion, this study elucidates the region-specific responses of porcine intestinal organoids to PEDV infection, demonstrating increased susceptibility and antiviral activation in jejunal and ileal organoids. These region-specific responses, driven by differential expression of viral entry receptors and immune-regulatory genes, advance our understanding of PEDV pathogenesis and suggest potential therapeutic targets for protecting neonatal piglets from PEDV.
















