INTRODUCTION
Insects are one of the most abundant organisms on the earth, and considering their biodiversity, they represent an abundant source of various bioactive compounds such as antimicrobial peptides (AMPs) [1,2]. AMPs derived from insects are known to exhibit anti-bacterial, anti-cancer, anti-fungi, anti-parasitic, anti-viral, and anti-inflammatory activities [3,4]. Among various AMPs, defensins, cecropins, drosomycins and attacins are currently being extensively investigated as alternatives to antibiotics [4]. In pig, AMPs have been studied as feed supplementation to alternate antibiotics, and they have shown beneficial effects on performance [5]. CopA3 (LLCIALRKK-NH2), derived from Copris tripartitus (i.e., the dung beetle), is an AMP with a similar structure to defensin and is known to exhibit anti-cancer, anti-bacterial, anti-inflammatory and anti-oxidant activities, as well as activation of innate immunity [6–10].
Muscle satellite cells (MSCs) are located between the myofiber plasmalemma and, along with the surrounding basal lamina, form new muscle fibers [11,12] through proliferation and differentiation upon activation by damage or physiological activities such as growth and exercise [13,14]. Muscle growth in mammals is accomplished via the addition of myonuclei which are supplied by MSCs, because myofibers nuclei cannot proliferate independently [15]. MSCs can increase the number of muscle fibers and are thus important factors for muscle growth [16]. In addition, research on the proliferation of MSCs can apply not only in skeletal muscle regeneration and disease [17], but also in the field of cultured meat research. The development of MSCs is precisely regulated by the expression of PAX7 and myogenic regulatory factors (MRFs) such as MYOD1, MYF5, MYOG, and MRF4 genes and [18–20]. In particular, the expressions of PAX7 and MYOD play important roles in the activation, proliferation and differentiation of MSCs [21,22]. The increased expression of PAX7 and MYOD indicates that the cells are highly proliferating. Moreover, since differentiation is initiated when PAX7 is down-regulated, the expression of PAX7 and MYOD is an important indicator for determining the proliferation and stemness of MSCs [20].
Since MSCs plays an important role in determining the rate of postnatal muscle growth [23], they are a key factor contributing to increased muscle mass. The growth performance of pig is closely related to their muscle growth, and these performances were found to be increased when pig was fed with AMP which was mixed with lactoferrin, cecropin, defensin, and plectasin [24]. However, the effects of CopA3 on porcine MSCs are yet to be explored in academic research. Here, we showed that CopA3 maintains the proliferative activity of MSCs by upregulating the expression levels of PAX7 and MYOD in MSCs.
MATERIALS AND METHODS
All animal experimental procedures in this study were approved by the Animal Ethics Committee of Jeonbuk National University, Korea (JBNU2019-020).
CopA3 (LLCIALRKK-NH2) was synthesized by AnyGen (Gwangju, Korea). The peptide was purified via high performance liquid chromatography (HPLC) using a Capcell Pak C18 column (Shiseido, Ginza, Japan). The peptide was then dissolved in distilled water and stored at −20°C until use.
MSCs were isolated from the femur skeletal muscle of 1-day-old male pig, following a protocol as described in our previous study [25]. The isolated MSCs were identification using flow cytometry Supplementary (Fig. S1). After isolation, MSCs cells were cultured in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12; Gibco, Carlsbad, CA, USA) with 15% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin-glutamine (PSG; Gibco) at a humidified atmosphere of 5% CO2. When the cells reached 90% confluency, MSCs were sub-cultured for increasing cells. MSCs were cultured in growth medium at a humidified atmosphere of 5% CO2. After 24 h, the medium was replaced with a culture solution supplemented with CopA3 at concentrations of 5, 10, and 25 μg/mL for 48 h. Control groups were grown without CopA3, and the medium was changed every 24 h.
Cell viability of MSCs under various CopA3 concentrations was analyzed with a cell counting kit-8 (CCK-8, Dojindo, Rockville, MD, USA) to select the optimal CopA3 concentration. MSCs were seeded in 96 well plates (5 × 103/well) and cultured in the growth medium in a humidified incubator with 5% CO2 at 37°C. After 24 h, MSCs were treated with various concentrations of CopA3 (i.e., 5, 10, 25, and 50 μg/mL) and without CopA3 as control for 48 h each. Each group of cells was set with five replicate wells. The CCK-8 solution was added to each well, and plates were incubated at 37°C and 5% CO2 for 4 h. Absorbance was determined using a microplate reader (Thermo Fisher Scientific, Rochester, NY, USA) at 450 nm.
MSCs were seeded in 6 well plate with 1.5 × 105 cells per well. All control and treatment groups were tested thrice. After 72 h, cells were detached using 0.25% Trypsin-EDTA (T.E) and neutralized with a washing medium (DMEM/F12 with 10% FBS and 1% PSG). Each group of cells was counted using a hemocytometer under an inverted microscope.
For flow cytometric analyses, MSCs of control and treatment groups (5, 10, and 25 μg/mL of CopA3) were cultured for 72 h and harvested using 0.25% T.E., before further washing with cold phosphate buffered saline (PBS) containing 1% bovine serum albumin (BSA; Sigma Aldrich, St. Louis, MO, USA) in Eppendorf (EP) tubes. For cell cycle analyses, samples were centrifuged using EP tubes at 200×g for 5 min at 4°C. The supernatant was discarded and then gently resuspended in 1 mL 70% ethanol. EP tubes were incubated for 5 min at 4°C and centrifuged at 850×g for 5 min at 4°C. After discarding the supernatant, we washed the cells twice with cold PBS containing 1% BSA. Cells were then stained with propidium iodide (PI; Bio Legend, San Diego, CA, USA) containing 100 μg/mL of RNase (Bio Basic Canada, Ontario, Canada). Apoptosis was determined using an fluorescein isothiocyana (FITC) Annexin Ⅴ Apoptosis detection kit with PI (Bio Legend), according to the manufacturer’s instructions. All samples were analyzed via fluorescence activated cell sorting (FACS) calibur flow cytometry (Becton, Dickinson Company, San Diego, CA, USA) and BD Cell Quest Pro software. A total of 10,000 events were collected per sample to manually determine the percentage of G1, S and G2/M phases.
MSCs were seeded on confocal dishes with 1 × 105 cells per well for immunocytochemistry assay. After 3 days, cultured MSCs were fixed with 4% cold paraformaldehyde in PBS for 20 min at room temperature (RT) and then rinsed thrice using PBS. Cells were permeabilized and blocked with a blocking solution (i.e., PBS containing 0.3% Triton X-100 and 3% BSA) for 1 h at RT. Cells were washed with 0.3% Triton X- 100 in PBS three times and stained overnight at 4°C using antibodies against PAX7 (1:50, DSHB, Iowa, IA, USA) and MYOD (1:200, Proteintech, Rosemont, IL, USA). After washing, cells were incubated with Alexa Flour-488 (1:1000, Molecular probes, Eugene, OR, USA) and Alexa Flour-568 (1:1000, Molecular probes) conjugated secondary antibodies in a dark room at RT. Cells were incubated with 4’-6-diamidino-2-phenylindole (DAPI, 1:1000) to visualize the nuclei for 5 min at RT. Confocal images were acquired using a super resolution confocal laser scanning microscope (SR-CLSM, LSM 880, Carl Zeiss, Oberkochen, Germany) and analyzed using ZEN imaging software.
Total protein was extracted from MSCs using the radioimmunoprecipitation assay (RIPA) buffer (Biosesang, Sungnam, Korea) containing protease inhibitor (Thermo Fisher Scientific) on ice for 40 min. After centrifugation for 30 min at 21,000×g, the supernatant was collected, and the protein concentration of cell lysates was measured with the detergent compatible (DC) protein assay kit (Bio-Rad, Hercules, CA, USA). Proteins were electrophoresed via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 12% acrylamide gel and transferred onto polyvinylidene fluoride (PVDF) membranes. Membranes were blocked using 5% skim milk in Tris buffer solution (TBS) containing 0.5% Tween 20 (TBST) for 1.5 h at RT, after which they were rinsed with TBST and incubated overnight at 4°C with the following primary antibodies: GAPDH (1:5000, Invitrogen, Carlsbad, CA, USA), PAX7 (1:1000, DSHB, Iowa, USA), MYOD (1:1000, Proteintech), MYOG (1: 1000, Abcam, Cambridge, UK). Membranes were rinsed with TBST, and incubated with secondary antibodies for 1.5 h at RT. Horseradish peroxidase (HRP)-conjugated secondary antibodies were goat anti-mouse IgG (1:2000-1:7500, Thermo Fisher, San Jose, CA, USA) and goat anti-rabbit IgG (1:2000, Thermo Fisher), which were used appropriately against primary antibodies. After washing with TBST, immunoblots were visualized using an enhanced chemiluminescene kit (Thermo Fisher), and the images were acquired with iBright CL 100 Imaging system (Thermo Fisher). All proteins were normalized with GAPDH.
Cells were collected and stored at −80°C. Total mRNA was isolated using RNeasy Mini kit (Qiagen, Valencia, CA, USA), according to manufacturer’s instructions. Total mRNA concentrations were quantified using a Nanodrop spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). The cDNA synthesis was performed using a cDNA synthesis kit (Bioneer, Daejeon, Korea). A primer, 1 μl of cDNA, and AMPIGENE® qPCR Green Mix (Enzo, San Diego, CA, USA) were prepared to a total volume of 20 μL, following the manufacturer’s protocol. Primer sequences of GAPDH, PAX7, MYOD1, and MYOG are listed in Table 1. Real-time quantitative PCR was performed using a CFX96TM Real-Time PCR detection system (Bio-Rad) in triplicate. All data were normalized with GAPDH and calculated using the 2-ΔΔCT method [26].
Data were analyzed using SAS version 9.4 (SAS Institute, Cary, NC, USA). One-way analysis of variance (ANOVA) was followed by Duncan’s Multiple Range Test to compare statistically significant differences in each group. All data are presented as mean ± SE, and statistical significance was set at p < 0.01 and p < 0.05.
RESULTS
We treated CopA3 (5, 10, 25, and 50 μg/mL) in MSCs to measure the viability and determined the optimal concentration (Fig. 1A). The 5 μg/mL group presented the highest cell viability. The viability was gradually decreased as the concentration of CopA3 increased (p < 0.01). In addition, cell viabilities of the 10 and 25 μg/mL groups were not significantly different from that of the control group. However, cell viability of the 50 μg/mL group reduced significantly (p < 0.01) compared to that of the control group.
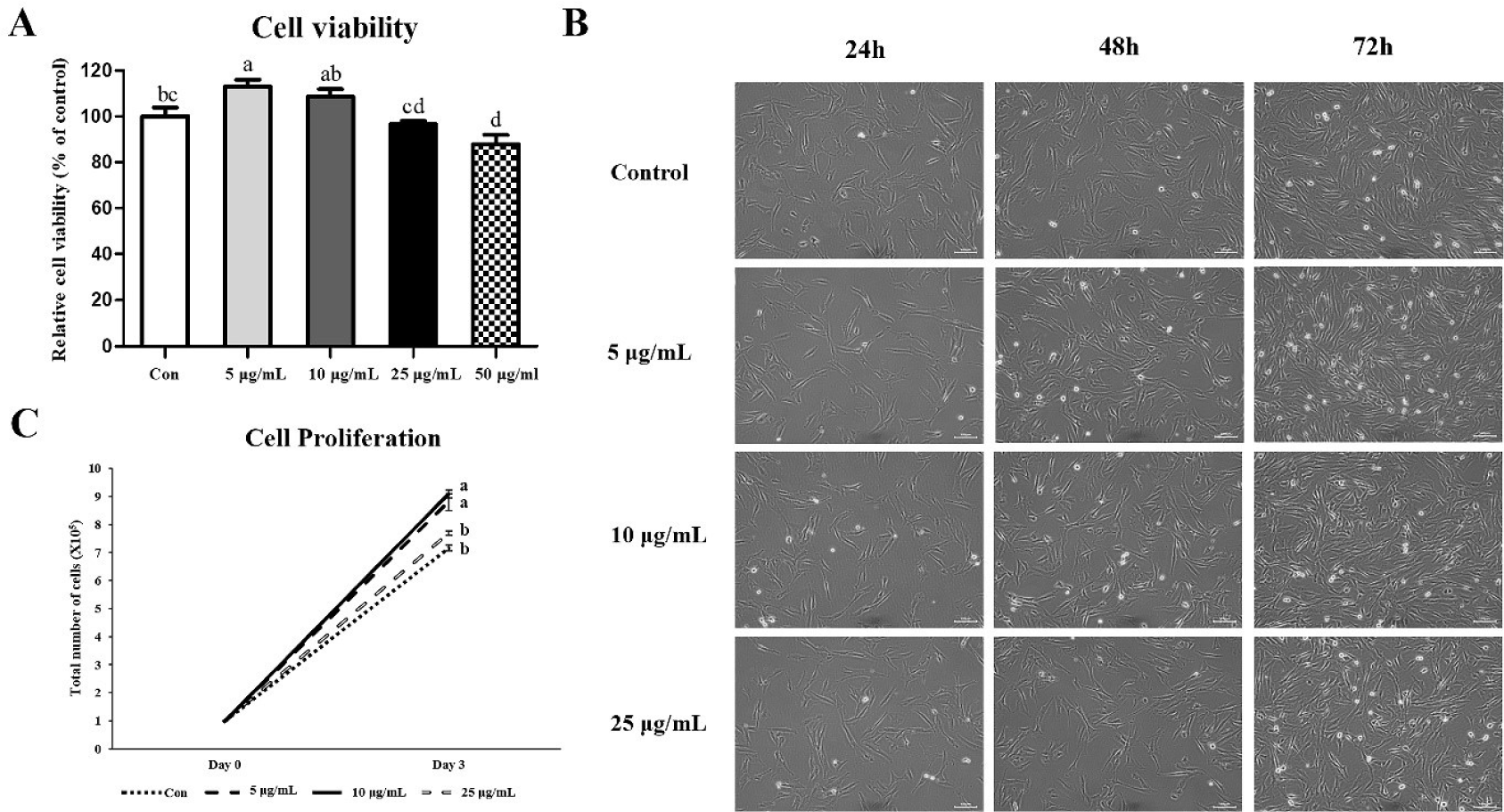
Cell morphology did not differ between control and treatment groups (i.e., 5, 10, and 25 μg/mL of CopA3) before treatment with CopA3 (Fig. 1B). However, differences in cell proliferation rates among the groups were observed starting 24 h after CopA3 treatment (48 h). On the third day of culture, we observed a higher number of cells in the 5 and 10 μg/mL groups than in 25 μg/mL and control groups (Fig. 1B). The proliferation rate of MSCs significantly increased (p < 0.01) in the 5 and 10 μg/mL groups compared to that in other groups, and the MSC proliferation rate of the 25 μg/mL group was not significantly different from that of the control group (Fig. 1C).
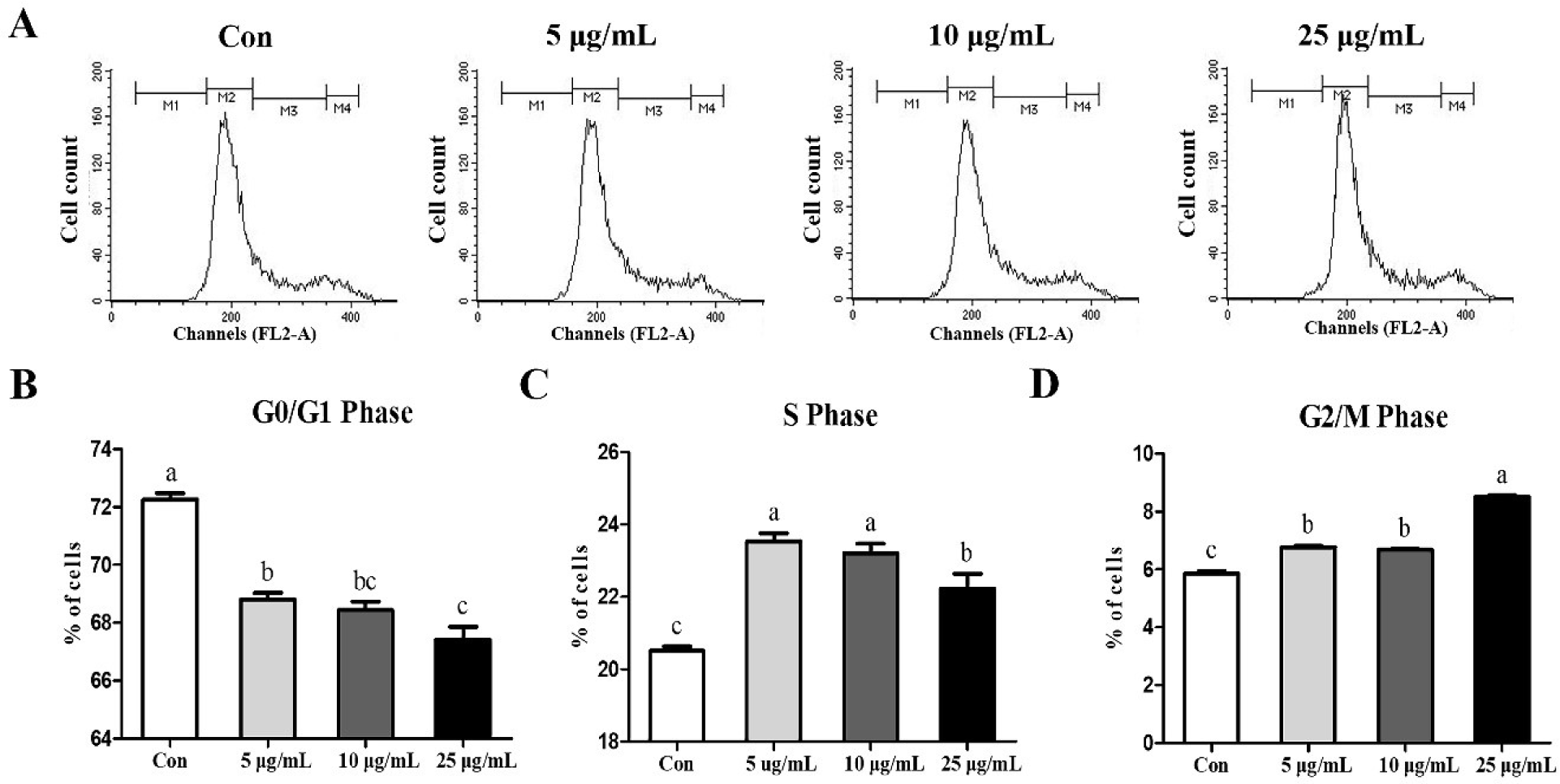
Cell cycle was analyzed via flow cytometry to determine whether the treatment of CopA3 affected the cell cycle of MSCs (Fig. 2A). CopA3 significantly decreased (p < 0.01) the percentage of the G0/G1 phase (Fig. 2B). Furthermore, compared to the control group, the CopA3 treated groups showed significantly increased (p < 0.01) the S and G2/M phases (Figs. 2C and 2D). Among all the groups, the proportion of the S phase was significantly higher (p < 0.01) in the 5 and 10 μg/mL groups and the lowest in the control (Fig. 2C). Moreover, the G2/M phase ratio was found to be the highest (p < 0.01) in the 25 μg/mL group (Fig. 2D).
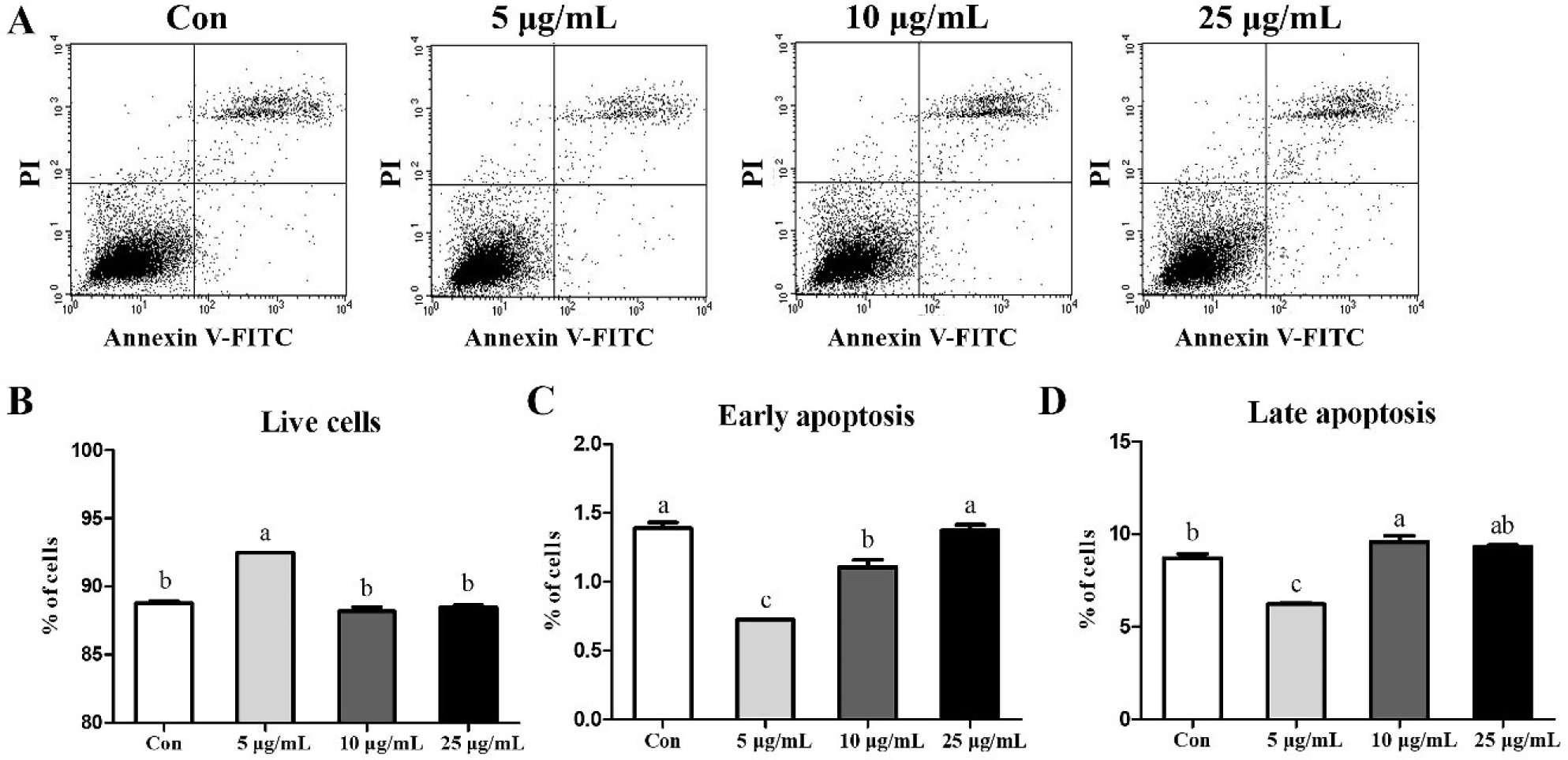
To determine whether CopA3 affects MSCs, we analyzed MSCs through FACS analyses (Fig. 3A). The results showed that treatment with 5 μg/mL CopA3 increased (p < 0.01) the number of live cells (Fig. 3B) and significantly decreased (p < 0.01) the number of early and late apoptotic cells (Figs. 3C and 3D). The number of early apoptotic cells in the 10 μg/mL groups was lower than that in the 25 μg/mL and control (Fig. 3C). However, the percentage of late apoptotic cells in 10 μg/mL groups were slightly increased (p < 0.01) than control group (Fig. 3D).
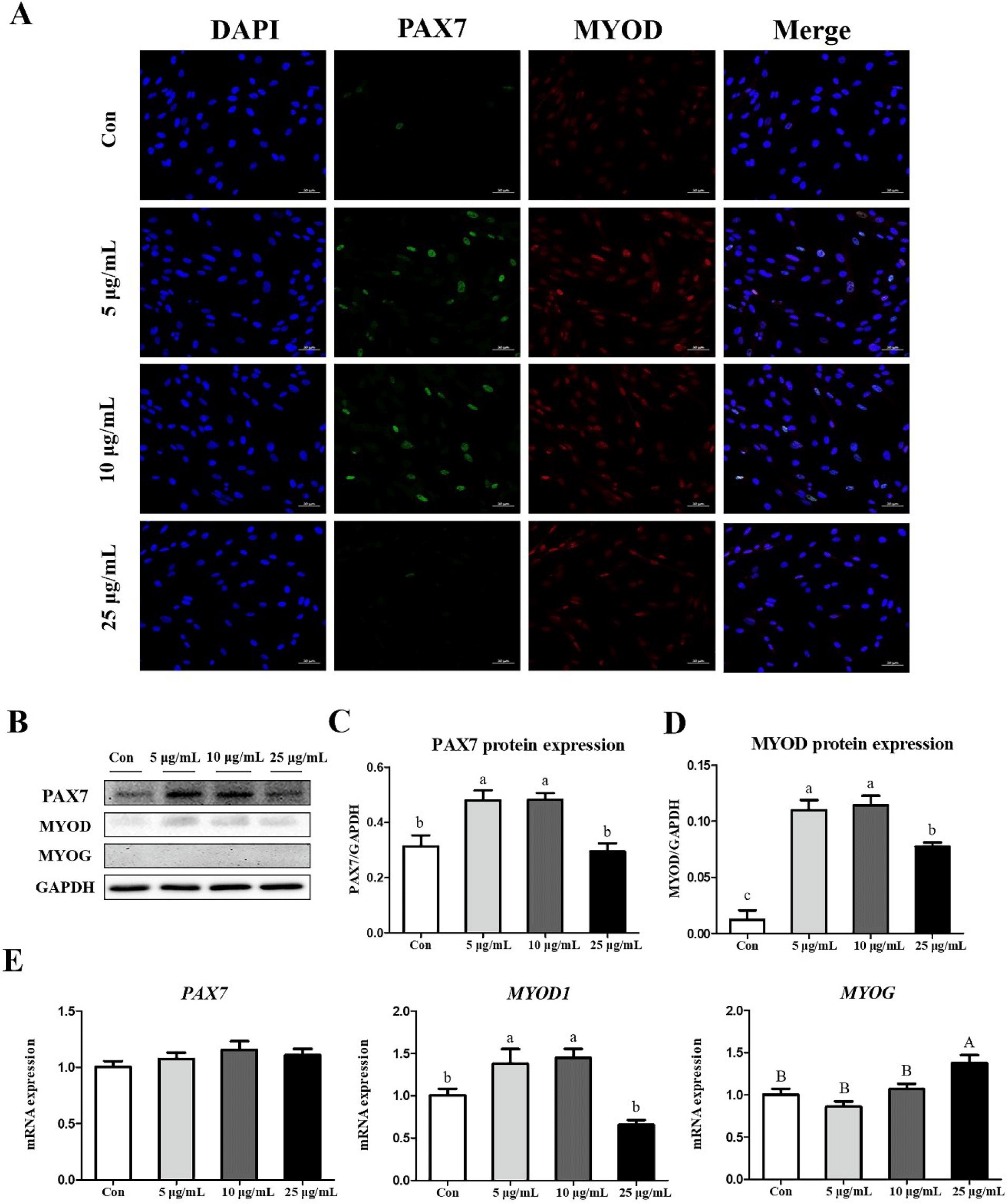
We also investigated the effects of CopA3 on myogenesis-related factors in MSCs. Results of the immunocytochemistry assays of PAX7 and MYOD showed that the expression of PAX7 was upregulated in the 5 and 10 μg/mL groups (Fig. 4A). In addition, results of western blot analyses were consistent with those of immunocytochemistry assays (Fig. 4B). The expressions of PAX7 and MYOD proteins were significantly upregulated (p < 0.01) in both 5 and 10 μg/mL groups (Figs. 4C and 4D), whereas the expression of MYOG was did not observed in any group (Fig. 4B). Results of mRNA expression analyses found that the expression level of MYOD1 was significantly higher (p < 0.01) in 5 and 10 μg/mL groups and that the highest (p < 0.05) expression of MYOG was observed in the 25 μg/mL (Fig. 4E). However, no significant differences in PAX7 expression were observed among all groups (Fig. 4E).
DISCUSSION
CopA3 regulates cell viability and proliferation, depending on the dose and cell type [8,27]. In this study, we showed that the addition of CopA3 to a growth medium at concentrations of 5 and 10 μg/mL increased both cell viability and the proliferation rate of MSCs. These results were similar to those of a previous study reporting the increased proliferation and cell viability of mouse neuronal stem cells [28] and human colonic epithelial cells [29] following 10 and 20 μg/mL treatment respectively. Some studies have reported that CopA3 selectively reduces the survival rate of cancer cell lines [10,27]; for example, when CopA3 was treated with 25 μg/mL in gastric cancer cells, cell viability decreased, and the proportion of necrosis and apoptosis increased [10]. In this study, treatment with 5 μg/mL CopA3 decreased the ratio of early and late apoptotic cells, and the addition of 25 μg/mL CopA3 did not increase the number of apoptotic cells. This could be due to the cell type-specific effects of CopA3.
The eukaryotic cell cycle is regulated by several types of cyclins, cyclin-dependent kinases (Cdks) and Cdk inhibitors (CKIs) [30,31]. p21 and p27 are Cip/kip family of CKIs, which negatively affect cell proliferation by inhibiting the cell cycle [32]. The increased expression of p21 and p27 in smooth muscle cells inhibits cell proliferation and induces growth arrest [33–35]. However, downregulation of p21 increased the proliferation of myoblasts [36]. Moreover, downregulation of p21 by basic fibroblast growth factor (FGF2) also increased the proliferation of muscle stem cells by promoting the transition from the G1 to S phase (i.e., G1/S transition) [37]. The rate of cell proliferation can be determined by the S phase cell ratio of the cell cycle [38]. The accelerated G1/S transition promotes the proliferation of MSCs in mice [39] and bovines [40]. In a previous study, CopA3 was found to increase the proportion of S and G2/M phases via the down-regulation of p27 expression in mouse neuronal stem cells [28]. CopA3 also changed ubiquitin ligase activity to downregulate p21 and increase the S phase of the cell cycle in epithelial cells [29]. In the present study, we demonstrated that the addition of CopA3 at final concentrations of 5 and 10μg/mL to the culture medium induced MSC proliferation by promoting the transition from G1 to S phase. However, the G2/M phase cell cycle arrest occurred when CopA3 was added at a final concentration of 25 μg/mL and indicated that cells cannot undergo mitosis due to DNA damage during the G2 phase [41].
Myogenic lineage is highly regulated by transcription factors such as PAX7, MYOD and MYOG [18,42]. PAX7 is expressed in all MSCs [18] and is an important transcription factor for the maintenance of MSCs, because it plays a role in returning activated MSCs to quiescence [43,44]. In addition, PAX7 induces proliferation of the myoblast, which is an activated state of MSCs [21,43]. Activation of MSCs induces the co-expression of PAX7 and MYOD [43], followed by a strong proliferation of myoblasts entering the cell cycle [45]. PAX7 expression upregulates MYOD and maintains a state of MSCs activation, but inhibits differentiation into myotube [46]. Subsequently, continuous differentiation signals induce the downregulation of PAX7 and increase the expression of MYOG to exit the cell cycle, resulting in the formation of new muscle fibers [45]. Therefore, PAX7(+)/MYOD(+)/MYOG(−) satellite cells exhibit active proliferation in an undifferentiated state [47]. In this study, the protein expressions of PAX7 and MYOD in MSCs were increased after treatments with 5 and 10 μg/mL of CopA3. In addition, no MYOG expression was observed. Therefore, we suggest that CopA3 might affect the maintenance of activated MSCs by upregulating PAX7 and MYOD expression.
In conclusion, our results suggest that CopA3 treatment at concentrations of 5 and 10 μg/mL increases MSCs proliferation by promoting the transition from G1 to S phase, as well as upregulating PAX7 and MYOD expression. Further studies are needed to identify the precise molecular mechanisms underlying these phenomena. This study may have implications for the application of insect-derived peptides in increasing the productivity of the livestock industry as well as the field of cultured meat.
SUPPLEMENTARY MATERIALS
Supplementary materials are only available online from: https://doi.org/10.5187/jast.2022.e81.
















