Lactic acid bacteria (LAB) have been associated with the fermentation and preservation of food since ancient times and are one of the most important groups of industrial microorganisms with a multi-billion dollar market. They are naturally found in fermented foods as well as in human and animal cavities, including the gastrointestinal tract [1,2]. LAB colonizes the intestines of animals as a part of the normal intestinal flora, inhibit the growth of harmful bacteria, prevent diarrhea, and inhibit the absorption of toxic substances [3]. Ruminants utilize intestinal microbes to break down cellulose into glucose so that it can be used as an energy source, and some LAB are known to use cellulose as a carbon source [4]. Ruminant roughage contains both fibrous (cell wall material) and non-fibrous (cell content) carbohydrates, but most of them are composed of fibers such as cellulose, hemicellulose, and pectin [5].
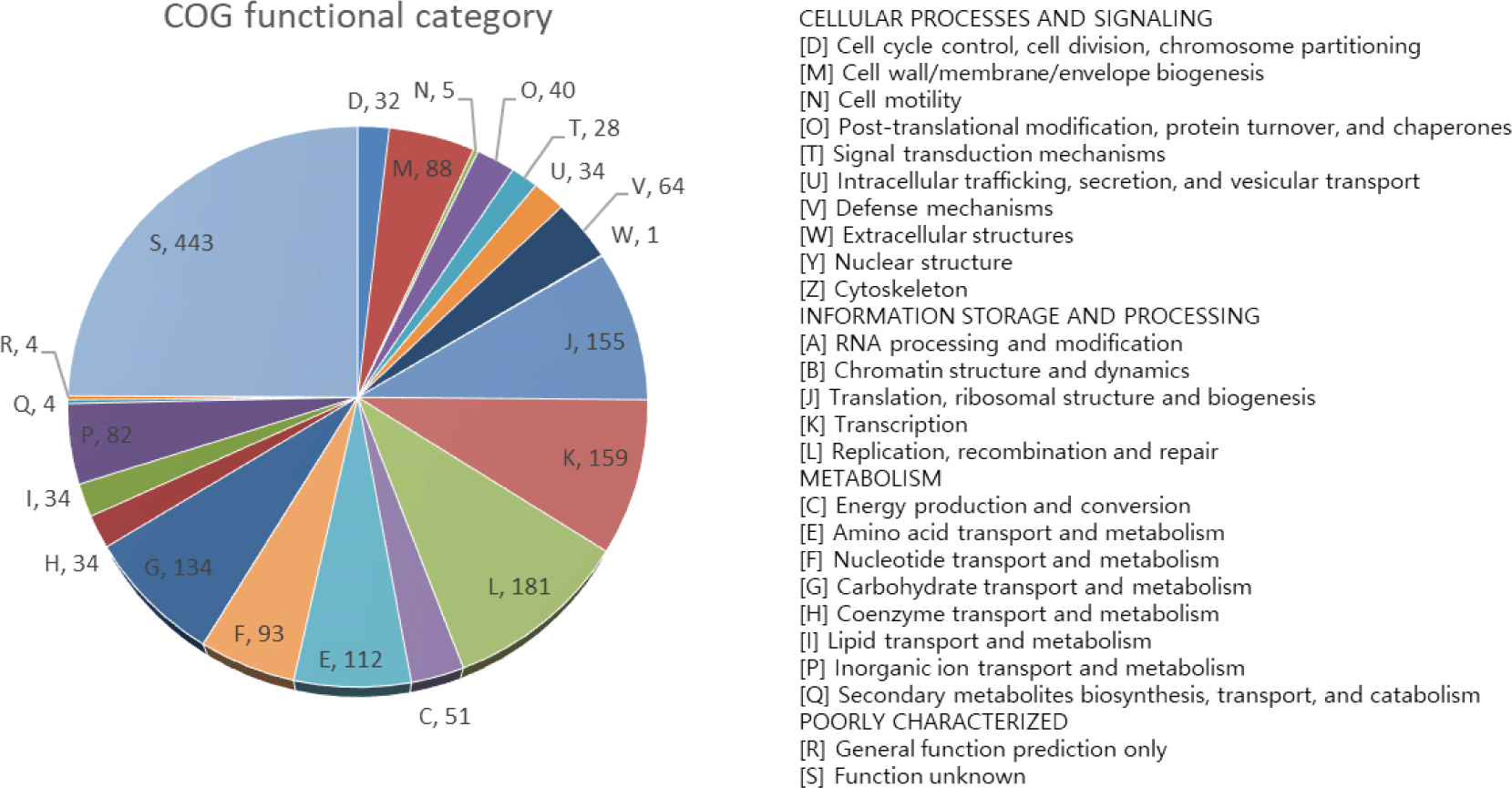
In the present study, Lactobacillus johnsonii 7409N31 (KCCM 13026P) was isolated from the feces of a healthy 11-day-old Hanwoo calf. Strain 7409N31 was anaerobically grown in deMan, Rogosa, and Sharpe (MRS, Difco, Franklin Lakes, NJ, USA) medium at 35°C for 24 h. Genomic DNA of 7409N31 strain was extracted as described previously [6]. The complete genome of L. johnsonii 7409N31 was sequenced by using Pacific Biosciences (PacBio) reagents (DNA Link, Seoul, Korea). A total number of 735,552 reads with a mean subread length of 5,638 bases (N50, 6,459 bases) were obtained with PacBio sequencing. These sequences were assembled de novo using the Hierarchical Genome Assembly Process (HGAP, version 3.0) workflow [7]. The genome of the L. johnsonii 7409N31 was annotated using the NCBI Prokaryotic Genome Annotation Pipeline and the Pathosystems Resource Integratin Center (PATRIC, version 3.6.12) genome data base [8]. Genome assembly completeness was evaluated using Benchmarking Universal Single-Copy Orthologous suite (BUSCO, version 5.1.3) [9], and evolutionary genealogy of genes: Non-supervised Orthologous Group-mapper (EggNOG-mapper, version 2.0, http://eggnog-mapper.embl.de) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (www.genome.jp/kegg) were used for functional annotation. The complete genome of strain 7409N31 has one circular chromosome (2,198,442 bp), with a guanine + cytosine (G + C) content of 35.01 mol%, 2,222 protein-coding sequences, 24 rRNA, 3 ncRNA, and 112 tRNA genes (Fig. 1 and Table 1). Further analysis showed that about 80% of the protein-coding genes (1,778 proteins) could be clustered in 21 functional categories of cluster of orthologous groups (COG) (Fig. 2). Most of the proteins of strain 7409N31 were classified into categories related to ‘core functions’ such as replication, recombination and repair (category L: 10.2%), transcription (category K: 8.9%), translation, ribosomal structure and biogenesis (category J: 8.7%), and carbohydrate transport and metabolism (category G: 7.5%). This suggests that strain 7409N31 has a relatively strong carbohydrate metabolism ability similar to other strains in the genus Lactobacillus [10].
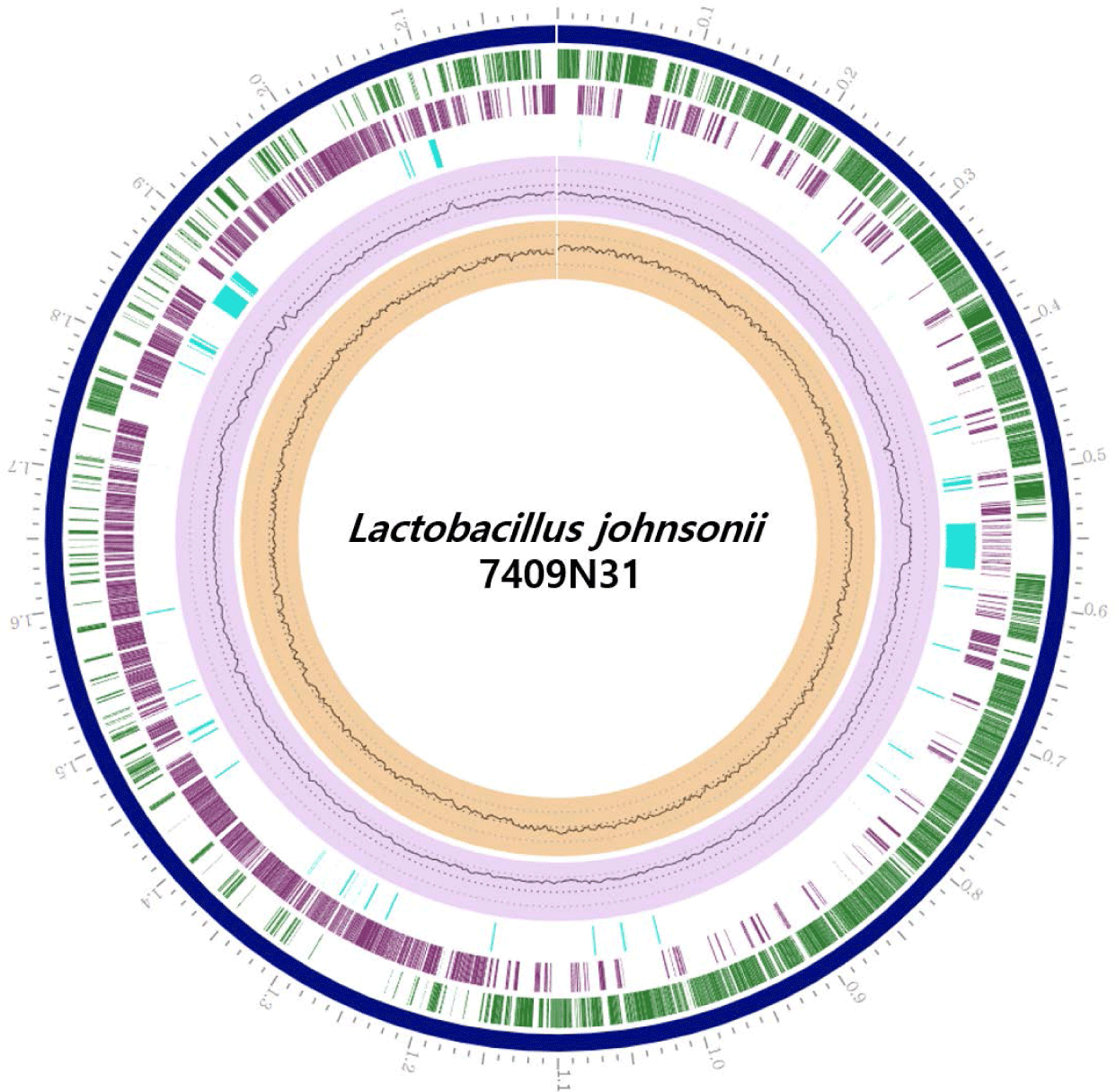
The genome of L. johnsonii 7409N31 possessed the bcsZ gene that encodes an endoglucanase involved in the hydrolysis of the D-glycosidic bond of cellulose. In addition, the presence of genes such as beta-fructofuranosidase (sacA), cellobiose PTS components (celB and chbC), and oligo-1,6-glucosidase (IMA, malL) involved in the metabolism of non-fibrous carbohydrates such as starch was confirmed. These results suggest that L. johnsonii strain 7409N31 has the potential to be developed as an industrial probiotic feed additive because it can facilitate digestion of both fibrous and non-fibrous carbohydrates.
















