RESEARCH ARTICLE
Complete genome sequence of Priestia megaterium S188, a hydrogen sulfide-degrading bacterium
Sang Hoon Kim1
,
Ji Hoon Song1
,
Remilyn M. Mendoza1
,
Dae-Kyung Kang1,*
Author Information & Copyright ▼
1Department of Animal Biotechnology, Dankook University, Cheonan 31116, Korea
*Corresponding author: Dae-Kyung Kang, Department of Animal Biotechnology, Dankook University, Cheonan 31116, Korea, Tel: +82-41-550-3655, E-mail:
dkkang@dankook.ac.kr
© Copyright 2025 Korean Society of Animal Science and Technology. This is an Open-Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received: Jul 08, 2024; Revised: Aug 17, 2024; Accepted: Aug 21, 2024
Published Online: May 31, 2025
Abstract
Priestia megaterium (formerly Bacillus megaterium) is a gram-positive, aerobic, spore-forming bacterium found in a wide range of environmental niches. Here, we report the complete genome sequence of P. megaterium S188 isolated from soil, which can decrease hydrogen sulfide (H2S) levels and help reduce malodor generation in livestock farms. Putative genes related to sulfide assimilation and conversion were found in the genome of P. megaterium S188; among these, one O-acetylhomoserine (O-AH) desulfhydrase, two cysteine synthases-primarily related to the biosynthesis of sulfur-containing amino acids, five rhodanese or sulfurtransferases, and one nitrogen reductase were identified. The genomic information on P. megaterium S188 provides insights into the possible biodegradation or conversion mechanisms of sulfur-containing substances that cause malodors, which can help reduce odor generation. Furthermore, identification of the key genes or molecules responsible for H2S reduction would facilitate the optimization of the H2S-degrading ability of S188.
Keywords: Priestia; Bacillus megaterium; Malodor; Hydrogen sulfide
Malodor generation during livestock production is a major problem in livestock farms, as it can negatively affect animals, humans, and the environment [1]. Particularly, hydrogen sulfide (H2S), which is a colorless gas heavier than air with a rotten egg-like odor, can cause severe distress to livestock workers [2,3]. Priestia megaterium S188, originally isolated from soil, can reduce H2S levels in manure [4]. In this study, we sequenced the complete genome of P. megaterium S188. Initially, S188 was grown in Nutrient Broth (Difco, Tucker, GA, USA) at 30°C for 24 h. The genomic DNA of strain S188 was extracted as described in a previous study [5], and its quality was checked using a spectrophotometer (UV-1601PC, Shimadzu, Kyoto, Japan). The genome of S188 was sequenced using the PacBio RSII platform (ver. 2.0; Pacific Biosciences) at Macrogen (Seoul, Korea). All the generated reads were de novo assembled using the RS HGAP Assembly (ver. 3.0) program. The assembled S188 genome was annotated using Prokka v.1.14.6, and the RAST was accessed at https://rast.nmpdr.org on May 10, 2024. BlastP (https://www.ncbi.nlm.nih.gov/blast/), UniProt (https://www.uniprot.org), ClustalOmega (https://www.ebi.ac.uk/jdispatcher/msa/clustalo), and EggNOG-mapper (http://eggnog-mapper.embl.de) were used for the annotation, alignment, and identification of proteins. KEGG (Kyoto Encyclopedia of Genes and Genomes) Mapper (https://www.kegg.jp/kegg/mapper/) was used to map the genes of strain S188 to different metabolic pathways, and the DNA plotter in Artemis (v.18.2) was used to generate the genome maps of strain S188.
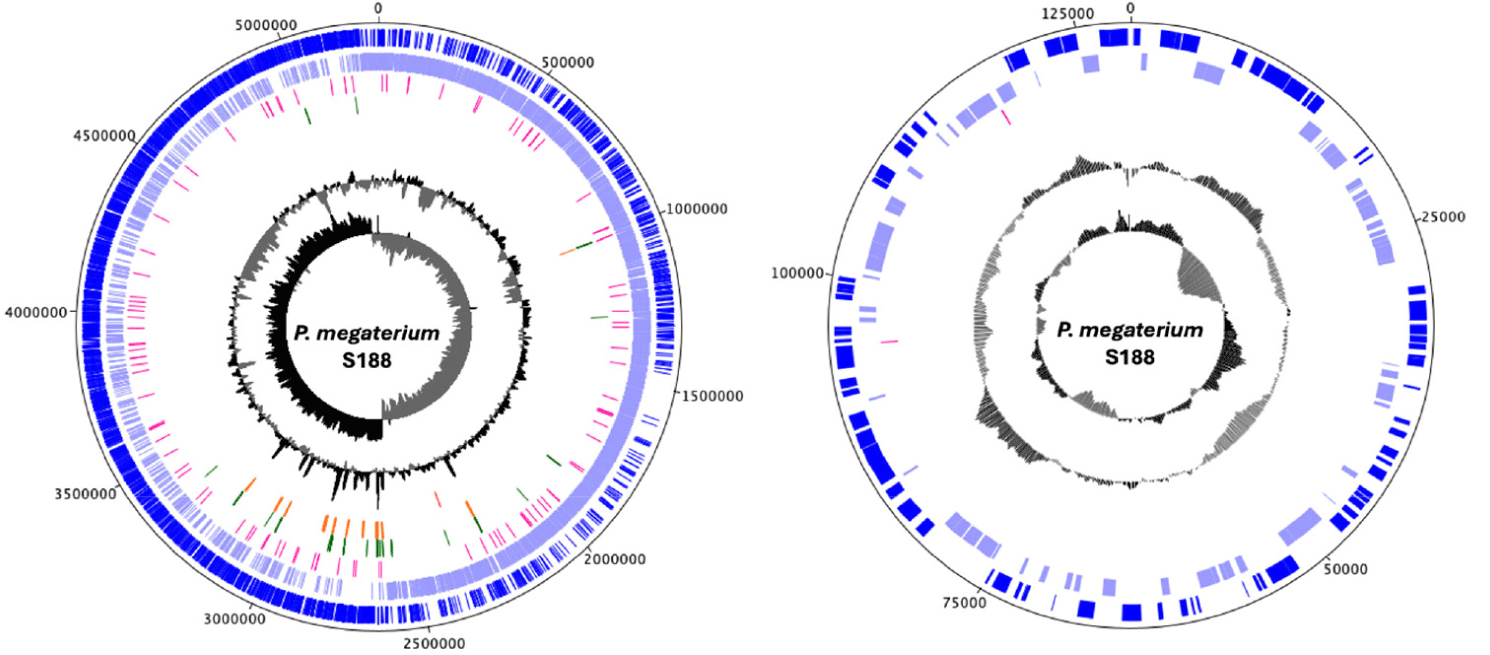
The S188 genome has a total length of 5,407,472 base pair (bp), with a chromosome size of 5,278,689 bp, and a putative plasmid of 128,783bp (Fig. 1). It has a guanine-cytosine (GC) content of 37.9% and comprises 5,761 genes, of which 5,494 are coding DNA sequences (CDS), 111 miscellaneous RNA, 118 transfer RNA, 37 ribosomal RNA, and one transfer-messenger RNA (Table 1). Genes related to H2S metabolism (i.e., assimilation/conversion) were identified using KEGG Mapper and manual curation of the reported genes associated with sulfur metabolism. Putative genes related to H2S assimilation and conversion, including one O-acetylhomoserine (O-AH) sulfhydrylase, two cysteine synthases, five rhodanese or sulfurtransferases, and one nitrogen reductase, were identified.
Fig. 1.
Genome maps of the Priestia megaterium S188 chromosome (Left) and plasmid (Right).
Circles illustrate the following features from the outside to the center: (1) Coding sequences on the forward strand, (2) coding sequences on the reverse strand, (3) Miscellaneous RNA (4) Transfer RNAs (tRNAs), (5) ribosomal RNAs (rRNAs), (6) G + C content, and (7) G + C skew. The figure was generated using DNA Plotter implemented in Artemis (v.18.2). G + C, guanine + cytosine.
Download Original Figure
Table 1.
Genome features of Priestia megaterium S188
| Features |
Chromosome |
Plasmid |
Total |
| Genome size (bp) |
5,278,689 |
128,783 |
5,407,472 |
| G + C content (%) |
38.0 |
34.2 |
37.95 |
| Total number of genes |
5,617 |
144 |
5,761 |
| Protein-coding genes |
5,352 |
142 |
5,494 |
| Misc RNA |
109 |
2 |
111 |
| tRNA genes |
118 |
0 |
118 |
| rRNA genes |
37 |
0 |
37 |
| tmRNA |
1 |
0 |
1 |
Download Excel Table
The amount of H2S released by yeast into the environment depends on the levels of sulfide (S2−) available [6]. Sulfide can be incorporated into sulfur-containing amino acids such as cysteine and methionine when it condenses with O-AH, a reaction catalyzed by O-AH sulfhydrylase, or with O-acetyl-L-serine, a reaction catalyzed by cysteine synthase [6,7]. Serine serves as a precursor for the biosynthesis of S-containing amino acids [6]; the expression of genes related to serine biosynthesis is lower in Saccharomyces. cerevisiae strains that produce H2S than in non-H2S producers. This suggests that the facilitation of S2− incorporation into amino acids due to increased amounts of intracellular serine results in reduced H2S production. [6]. In the S188 genome, genes for the assimilation of sulfide as well as the biosynthesis of serine, cysteine, and methionine were mapped using KEGG Mapper (Table 2).
Table 2.
Genes related to sulfide assimilation and conversion identified in the genome of Priestia megaterium S188
| Gene group |
Locus Tag |
Gene product |
| Sulfide assimilation |
S188_ch_03715 |
O-acetyl-L-homoserine sulfhydrylase |
| S188_ch_05019 |
Homoserine O-acetyltransferase |
| S188_ch_02391; S188_ch_02939 |
Cysteine synthase |
| Serine biosynthesis |
S188_01905 |
D-3-phosphoglycerate dehydrogenase |
| S188_ch_03473 |
Phosphoserine aminotransferase |
| S188_ch_03080; S188_ch_03084 |
Phosphoserine phosphatase |
| S188_ch_03845 |
Putative phosphoserine phosphatase 2 |
| Cysteine biosynthesis |
S188_ch_02414 |
S-adenosylmethionine synthase |
| S188_ch_03749 |
Homocysteine S-methyltransferase |
| S188_ch_02138 |
5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase |
| S188_ch_02426 |
S-ribosylhomocysteine lyase |
| S188_ch_02137 |
O-acetylserine-dependent cystathionine beta-synthase |
| S188_ch_02136; S188_ch_04308 |
Cystathionine gamma-lyase |
| Methionine biosynthesis |
S188_ch_01671; S188_ch_02273; S188_ch_05176 |
Aspartokinase |
| S188_ch_01672 |
Aspartate-semialdehyde dehydrogenase |
| S188_ch_02530 |
Homoserine dehydrogenase |
| S188_ch_03476 |
O-acetyltransferase |
| S188_ch_02505; S188_ch_043091 |
Cystathionine beta-lyase |
| S188_ch_04180 |
Methionine synthase |
| Rhodanese/sulfurtransferases |
S188_ch_00937 |
Putative rhodanese-like domain-containing protein |
| S188_ch_02024 |
Thiosulfate sulfurtransferase |
| S188_ch_02398 |
Sulfurtransferase |
| S188_ch_05007 |
Putative thiosulfate sulfurtransferase |
| S188_ch_05516 |
Rhodanese-related sulfurtransferase |
| Nitrogen reduction |
S188_ch_03676 |
Nitrate reductase |
Download Excel Table
In eukaryotic cells, rhodanese, a mitochondrial sulfur transferase, is part of the mitochondrial sulfide oxidation pathway involving sulfide quinone oxidoreductase, persulfide dioxygenase (PDO), and sulfite oxidase. This pathway ultimately oxidizes H2S to thiosulfate and sulfate [8]. In some bacteria, particularly Staphylococcus aureus, naturally occurring PDO-rhodanese fusion proteins (i.e., CstB) are involved in H2S detoxification [8,9]. Five putative sulfur transferases or rhodanese-like domain-containing proteins, which showed 18%–––27% similarity to the CstB of S. aureus, were also identified in the genome of S188 (Table 2).
The presence of nitrate reductase (Table 2) in the S188 genome indicates another possible mechanism whereby S188 can remove or reduce H2S. H2S removal using nitrate-reducing and sulfide-oxidizing bacteria has also been explored; herein, H2S serves as the electron donor for the reduction of nitrate to nitrogen gas [10].
P. megaterium S188 isolated from the soil can reduce the levels of H2S in manure and has the potential to reduce malodors in livestock farms. The involvement of the identified putative genes related to this phenotype, such as O-AH sulfhydrylase, cysteine synthase, putative sulfur transferases, rhodanese-like domain-containing proteins, and nitrate reductase, warrants further experimentation and validation. Nonetheless, the identification of these genes offers insights into the possible mechanisms by which S188, whether alone or in synergy with other nitrate-reducing, sulfur-oxidizing bacteria, reduces the levels of H2S and would facilitate the optimization of S188 activity to achieve more efficient H2S removal. Finally, complete cobalamin biosynthetic (cob) operon was also deteced in the genome of P. megaterium S188 (data not shown), indicating that S188 is able to synthesize vitamin B12, which needs to be investigated in the future.
NUCLEOTIDE SEQUENCE ACCESSION NUMBER
The genome sequences of P. megaterium S188 are available at GenBank with the accession number NZ_CP049296.1.
Acknowledgements
We would like to acknowledge the authors and developers of the tools that we used in analyzing the genome of Priestia megaterium S188 which we failed to cite in the paper due to the limitation in the number of citations.
Availability of data and material
Ethics approval and consent to participate
REFERENCES
Hong SH, Lee EY. Study on the reduction of livestock malodor using microbial agents-focusing on swine facilities. J Odor Indoor Environ. 2018; 17:85-94


Kang DK, Kim SH, Oh JK. inventor Dankook University Cheonan Campus Industry-Academic Cooperation Group, assignee. Bacillus megaterium S188 strain having enzyme secretion activity and hydrogen sulfide odor removal activity and uses thereof. Korean patent KR102196585B1 2020 Dec; 30