Bacteriocin production in lactic acid bacteria (LAB) has been consistently gaining attention owing to its potential as a viable alternative to antibiotics. Bacteriocins are ribosomally-synthesized peptides secreted by the producing strain. These peptides can have either a narrow or broad spectrum of activity, which indirectly determines the niche of the producing strain. The production of bacteriocins is generally viewed as a positive trait as it enables the producing strain to hinder potential competitors in the immediate environment as well as inhibit potentially harmful microorganisms [1]. The proteinaceous nature of bacteriocins renders them suitable for human use as they can be inactivated by digestive proteases. With the rapid development of antimicrobial drug resistance in microorganisms [2], research efforts focused on developing alternative solutions must be prioritized.
Commonly associated with vertebrate hosts, Ligilactobacillus is a genus of lactic acid bacteria composed of members that are homofermentative, non-motile, and urease-positive. Their ability to survive in gastric acid conditions and their Qualified Presumption of Safety (QPS) status from the European Food Safety Authority (EFSA) [3] make them popular choices for probiotics. Furthermore, the production of various antimicrobial salivaricins among strains of Lig. salivarius is well accounted for the development of probiotic strains [4]. In the present study, we report the genome analysis of a bacteriocin-producing Ligilactobacillus salivarius (formerly Lactobacillus salivarius) strain B4311, which was isolated from fecal samples collected from broiler chickens.
Strain B4311 was routinely cultured in de Mann, Rogosa, Sharpe ([MRS] BD Difco, Franklin Lakes, NJ, USA) broth supplemented with 0.05% L-cysteine, and incubated aerobically at 37°C for 24 h. Genomic DNA was extracted using the MagAttract HWM DNA Kit (Qiagen, Hilden, Germany) and quantified using Qubit ds DNA HS assay kit (Invitrogen, Waltham, MA, USA) with the Epoch™ Spectrometer (BioTek, Winooski, VT, USA). The genome was sequenced using the Pacific Biosciences (PacBio, Menlo Park, CA, USA) Sequel II platform. De novo assembly of the sequence reads was performed using the PacBio SMAR Analysis program (ver. 2.3.0). Functional annotation of the genome was performed using PRODIGAL ver. 2.6.2 [5] software and compared against protein databases (SwissProt, KEGG, SEED, EggNOG). Rapid annotation was employed using Subsystem Technology (RAST) with default parameters (https://rast.nmpdr.org/). Transfer RNAs (tRNA) and non-coding ribosomal RNAs (rRNA) were identified using tRNAscan-SE ver. 1.3.1 [6] and INFERNAL ver. 1.1.3 [7], respectively.
The complete genome of Lig. salivarius B4311 is 2,071,255 base pair (bp) which is assembled into three contigs: a single chromosome (1,801,655 bp), one megaplasmid (247,930 bp), and a small plasmid (21,670 bp) with a guanine + cytosine (GC) content of 33.1%. In addition, the genome contains 1,963 protein-coding sequences (CDS), 22 rRNA genes, and 78 tRNA genes. The genome features and circular maps of strain B4311 are presented in Table 1 and Fig. 1, respectively. Antimicrobial resistance genes, specifically for tetracycline and glycopeptides, were also detected via Resistance Gene Identifier ([RGI] https://card.mcmaster.ca/home). Among the 1,963 CDS, 1,241 were predicted with biological functions associated with cell cycle (n = 23), cell wall and motility (n = 116), cellular response (n = 69), DNA processing (n = 154), RNA processing (n = 119), protein processing (n = 202), defense mechanism (n = 31), energy production (n = 63), and transport and metabolism (n = 464). Additionally, 61 putative genes were detected with putative functions including stress response, DNA and RNA processing, antibiotic resistance, periplasm signaling, acetylation, amino acid transport, and production of enzymes including various hydrolases, methyltransferases, and transport proteins.
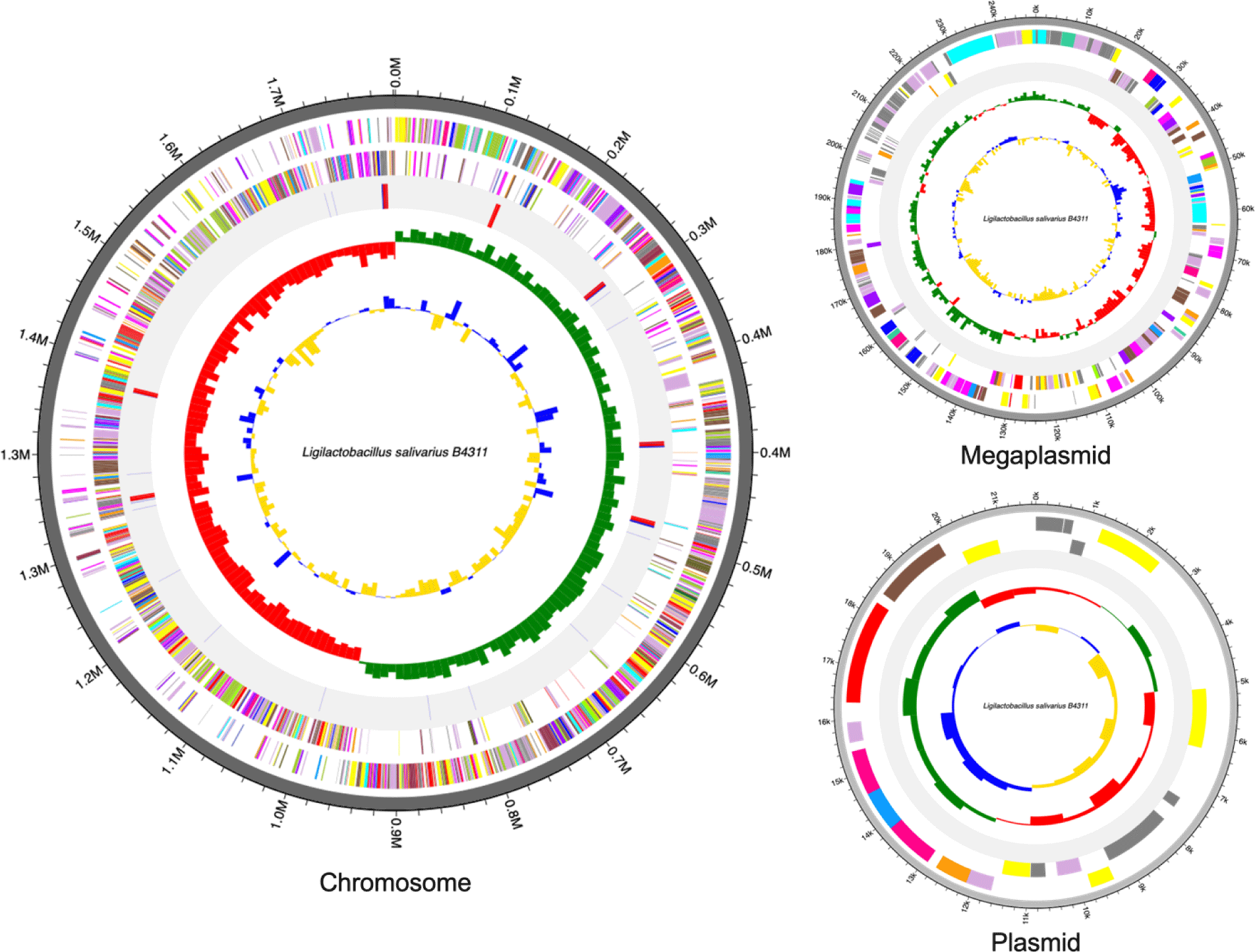
In silico analysis of the B4311 genome using BAGEL4 online program (http://bagel4.molgenrug.nl/) revealed the presence of a bacteriocin gene cluster for salivaricin P, a family of two-peptide bacteriocins belonging to class IIb. This bacteriocin family was originally discovered from Lig. salivarius DPC6005 [6] and is commonly produced among strains of Lig. salivarius isolated from animals intestines [8]. The salivaricin P gene cluster is located in the repA-type megaplasmid. Although the presence of megaplasmids is considered a typical feature of Lig. salivarius, variations exist among megaplasmid-encoded traits, including contingency metabolism genes (i.e., assimilation of sugars) and the presence or absence of bacteriocin genes, which provides a competitive advantage.
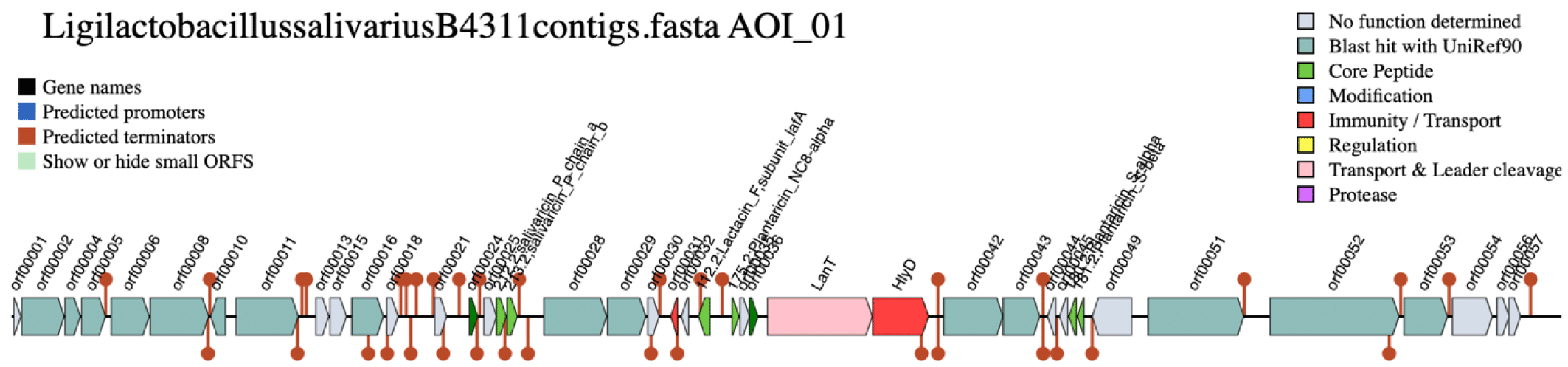
The genetic architecture of the bacteriocin gene cluster (Fig. 2) revealed the presence of two open reading frames (ORFs) encoding the salivaricin P chain A and chain B. The two peptide chains share a homologous sequence. Located downstream of the genes for the bacteriocin peptides are two ORFs encoding a histidine kinase and AbpR, which function as regulator proteins [9]. These are followed by AbpIM which encodes an immunity protein. These five ORFs are flanked by two comC genes, which have been reported as competence-stimulating peptide precursors in streptococci [10]. At the 3’ end of the gene cluster, two export proteins, LanT and HlyD were detected, encoding AbpT and AbpD bacteriocin export accessory proteins, respectively. Several ORFs encoding bacteriocin core peptides (i.e., lactacin F and plantaricins) were also detected. However, the similarity of these genes with the reference was poor, suggesting that the translated peptides might be inactive.
Production of active bacteriocins was demonstrated by spot-on-lawn assay against Listeria monocytogenes ATCC 19114 and ATCC 19115 strains. The cell-free supernatant of strain B4311 successfully inhibited the growth of Lis. monocytogenes (unpublished data), a common foodborne pathogen associated with raw and unpasteurized milk and the causative agent of listeriosis. The genomic information presented in this study confirms the ability of strain B4311 to elaborate bioactive peptides, which can have valuable applications in the food and animal industries.
















