Postbiotics are bioactive cellular components that are not classified as probiotics, prebiotics, or paraprobiotics, and may contain purified or a mixture of soluble factors, metabolic products and/or by-products, and other cell components that confer a beneficial health effect on the host. Bacteriocins, defined as antimicrobial peptides synthesized by the ribosome, are considered postbiotics that may have beneficial effects on the host, directly or indirectly [1]. The proteinaceous nature of these substances makes them susceptible to hydrolysis by endogenous proteolytic enzymes from animals or humans and exerts antibacterial, antibiofilm, or potentially anti-cancer properties [2]. Thus, bacteriocins are becoming increasingly important in the dairy and feed sectors for biopreservation and as substitutes for antibiotics. In contrast, ISAPP defined probiotics as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” [3]. Although probiotics are generally regarded as safe (GRAS), there is still an imminent risk of transmission of harmful genes such as antimicrobial resistance and virulence factor genes. Meanwhile, postbiotics offer several benefits, such as safer delivery, extended shelf life, and less risk of acquiring and spreading resistance genes and other harmful factors [4].
Different classes of bacteriocins include Class I and Class II bacteriocins, consisting of small molecular-size (≤ 10 kD), heat-stable bacteriocins, and Class III bacteriocins, comprised of small, heat-labile bacteriocins. Class I is further divided into subclass Ia and Ib corresponding to ‘lantibiotics’ and ‘circular bacteriocins,’ while Class II is divided into subclass IIa to IId, corresponding to ‘pediocin-like bacteriocins’, ‘two-peptide bacteriocins’, ‘leaderless bacteriocins’ and ‘non-pediocin-like single peptide bacteriocins’, respectively. Lastly, Class III can either be ‘bacteriolysin bacteriocin’ or ‘non-lytic bacteriocin’ [4]. The extensive range of bacteriocins provides prospects for investigating alternatives to traditional antimicrobials and requires thorough research to accurately define and apply these bioactive peptides with great precision.
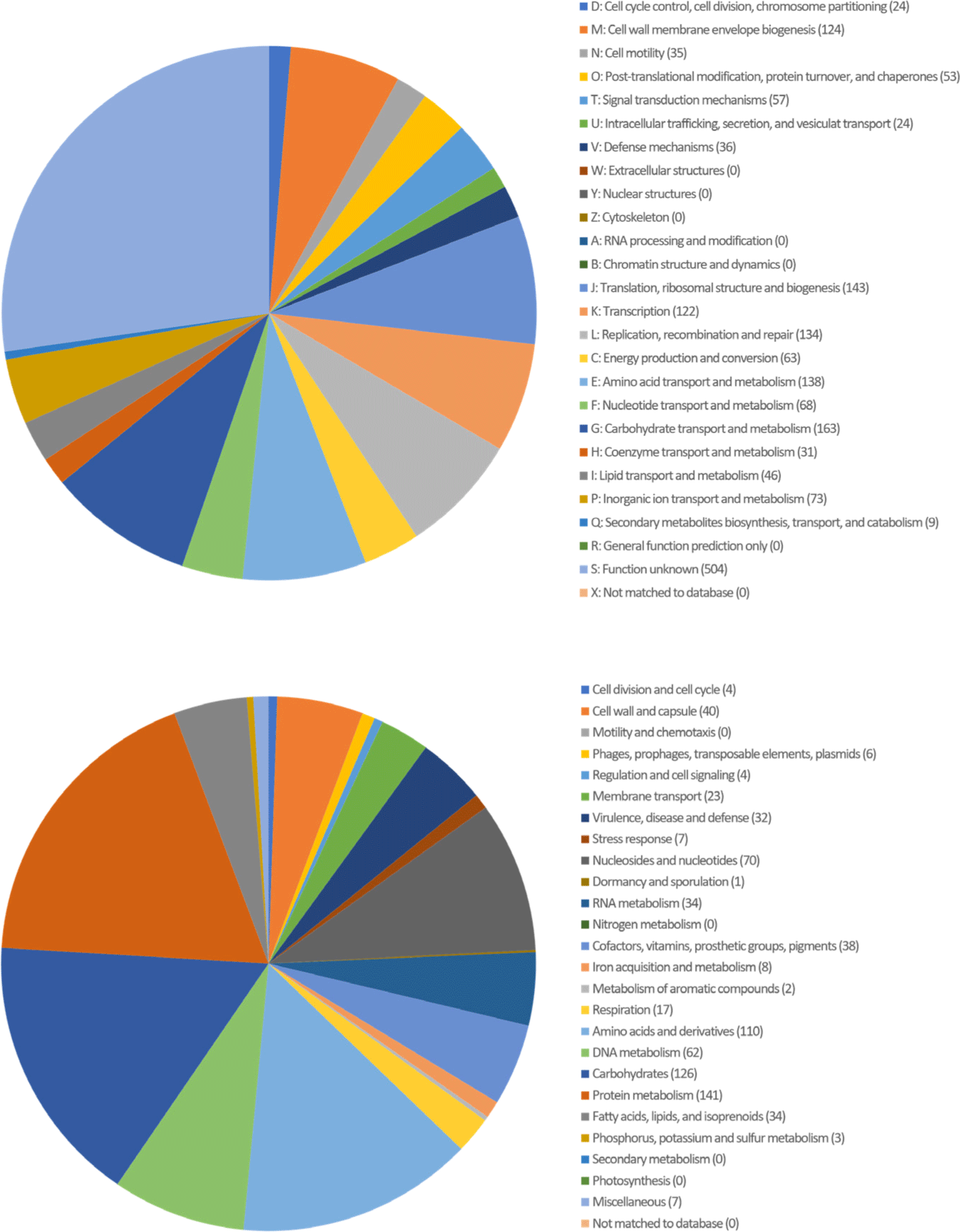
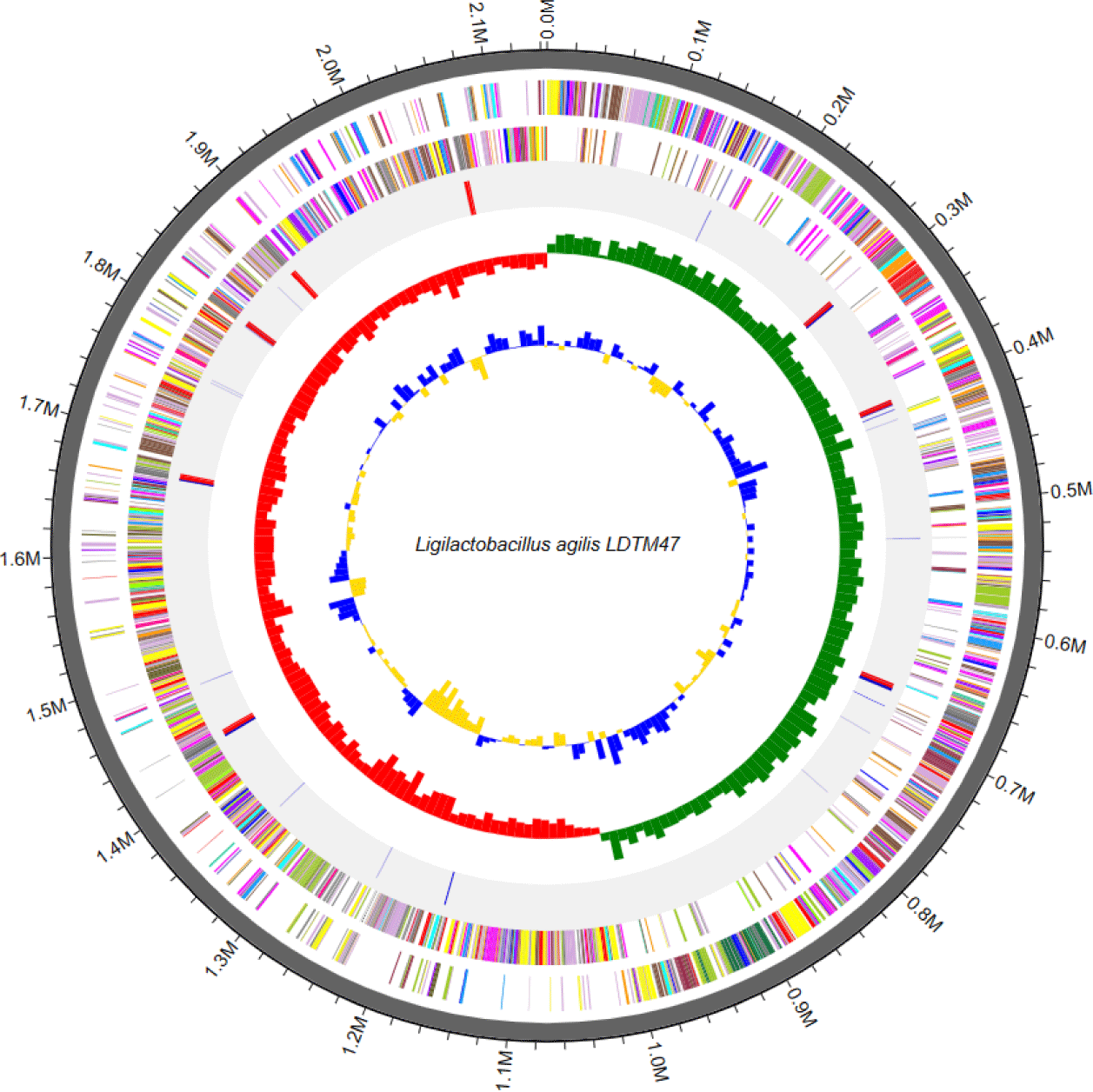
The bacteriocin-producing Ligilactobacillus agilis LDTM47 strain was isolated from the gastrointestinal tract contents (jejunum and ileum) of 5-week-old broilers from a farm affiliated with Chung-Ang University (Anseong, Korea). Lig. agilis LDTM47 is a Gram-positive, facultatively anaerobic, and rod-shaped bacteria. Most lactic acid bacteria are non-motile; however, Lig. agilis exerted motility and was later observed to be flagellated [5]. Generally, Lig. agilis LDTM47 was cultured aerobically in de Man, Rogosa, and Sharpe (MRS) medium (BD Bacto) at 37°C for 24 h [6]. The genomic DNA was sequenced using the Pacific Biosciences (PacBio, Menlo Park, CA, USA) RSII Single Molecule Real-Time (SMRT) platform and a 20-kb SMRKbellTM template library. The PacBio reads were assembled using the FALCON 0.5 program de novo. Functional categorization and annotation via Rapid Annotation using Subsystem Technology (RAST) (http://rast.nmpdr.org/) and CLgenomicsTM ver. 1.55 software and Cluster of Orthologous Groups (COG) derived from the EZBioCloud data were performed [4]. Functional annotation of protein-coding genes was performed using PRODIGAL ver. 2.6.2 software (Fig. 1) [7]. Putative bacteriocin genes were verified in silico using the BAGEL4 software (http://bagel4.molgenrug.nl/). The Lig. agilis LDTM47 whole genome sequencing (Fig. 2) showed a 2,144,466 base pair genome with a guanine + cytosine (GC) content of 41.9%. The genome was composed of a single contig with an N50 value of 2,144,466 bp. The genome comprises 2,131 protein-coding genes, 90 tRNA genes, and 24 rRNA genes, as shown in Table 1.
| Attribute | Value |
|---|---|
| Genome size (bp) | 2,144,466 |
| GC content (%) | 41.9 |
| No. of contigs | 1 |
| Total genes | 2,245 |
| Protein-coding gene | 2,131 |
| tRNA | 90 |
| rRNA | 24 |
| Plasmids | 0 |
| GenBank Accession No. | CP141636 |
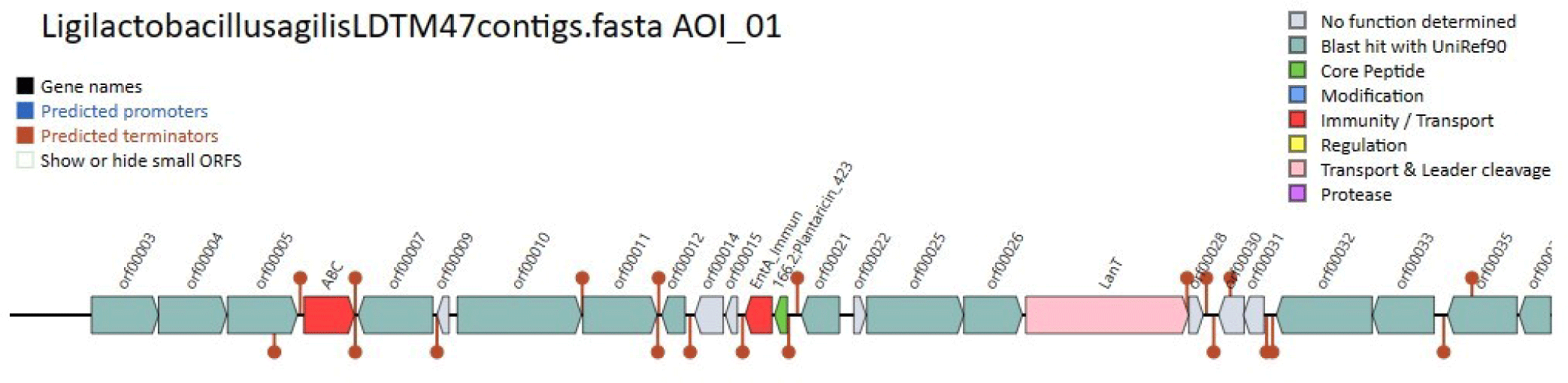
BAGEL4 analysis revealed that Lig. agilis LDTM47 harbors the core peptide gene, immunity, and transport genes for bacteriocin production (Fig. 3). One open reading frame (ORF) was predicted, encoding the bacteriocin core peptide with the amino acid sequence of MENKKK LTKADLAKVTGGSRYYGNGVTCGKHKCTVNWGQAWTCGVNRLANFGHGNC. The ‘YGNGV’ motif is associated with pediocin-like bacteriocin [8], suggesting that LDTM47 bacteriocin is a Class IIa bacteriocin. The lanT encodes the AbpT bacteriocin export accessory protein [9], and the abc encodes the import adenosine triphosphate (ATP)-binding protein FhuC [10]. Additionally, entA encodes the bacteriocin immunity protein. In silico characterization revealed that LDTM47 bacteriocin is stable with an instability index (II) of 1.32 (https://web.expasy.org/cgi-bin/protparam/protparam). Additionally, the bacteriocin was predicted to be susceptible to a number of proteolytic enzymes, including Arg-C proteinase, Asp-N endopeptidase, enterokinase, pepsin, proteinase K, and trypsin (https://web.expasy.org/cgi-bin/peptide_cutter/peptidecutter.pl). A BLASTp search of the LDTM47 amino acid sequence against L. agilis (taxid:1601) yielded only a limited number of significant alignments, indicating that the bacteriocin has received relatively little research interest thus far. Further, the sequence was searched in the RCSB Protein Data Bank and revealed the most relevant sequence identity (63%) with leucocin A, having 13 amino acid differences in (K20R, H27T, T29G, S31H, G32K, S34T, E39Q, F41W, S42T, A43C, H46C, G51N, and N53H). To our knowledge, only four Lig. agilis strains of chicken origin have been studied. Out of these strains, only one was found to produce a bacteriocin (garvicin), implying the need for further investigation on these bacteriocins.
Preliminary characterization of the physicochemical properties of LDTM47 bacteriocins revealed temperature and pH stability (data not shown) consistent with their Class IIa classification and in silico characterization of their stability, suggesting their safety and suitability in food and feed system applications. Although Lig. agilis LDTM47 strain lacks resistance to low pH and bile acids, rendering it challenging for probiotic development, its bacteriocin production may have potential applications as postbiotics, as biopreservation, and antibiotic alternatives.
















